Jmjd2/Kdm4 demethylases are required for expression of Il3ra and survival of acute myeloid leukemia cells
- PMID: 27257215
- PMCID: PMC4911927
- DOI: 10.1101/gad.280495.116
Jmjd2/Kdm4 demethylases are required for expression of Il3ra and survival of acute myeloid leukemia cells
Abstract
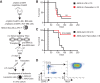
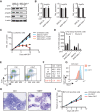
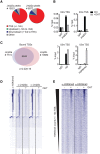
Acute myeloid leukemias (AMLs) with a rearrangement of the mixed-linage leukemia (MLL) gene are aggressive hematopoietic malignancies. Here, we explored the feasibility of using the H3K9- and H3K36-specific demethylases Jmjd2/Kdm4 as putative drug targets in MLL-AF9 translocated leukemia. Using Jmjd2a, Jmjd2b, and Jmjd2c conditional triple-knockout mice, we show that Jmjd2/Kdm4 activities are required for MLL-AF9 translocated AML in vivo and in vitro. We demonstrate that expression of the interleukin 3 receptor α (Il3ra also known as Cd123) subunit is dependent on Jmjd2/Kdm4 through a mechanism involving removal of H3K9me3 from the promoter of the Il3ra gene. Importantly, ectopic expression of Il3ra in Jmjd2/Kdm4 knockout cells alleviates the requirement of Jmjd2/Kdm4 for the survival of AML cells, showing that Il3ra is a critical downstream target of Jmjd2/Kdm4 in leukemia. These results suggest that the JMJD2/KDM4 proteins are promising drug targets for the treatment of AML.
Keywords: H3K9 methylation; JMJD2; acute myeloid leukemia; epigenetics; histone demethylase; interleukin 3.
© 2016 Agger et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- Bavetsias V, Lanigan RM, Ruda GF, Atrash B, McLaughlin MG, Tumber A, Mok NY, Le Bihan YV, Dempster S, Boxall KJ, et al. 2016. 8-substituted pyrido[3,4-d]pyrimidin-4(3H)-one derivatives as potent, cell permeable, KDM4 (JMJD2) and KDM5 (JARID1) histone lysine demethylase inhibitors. J Med Chem 59: 1388–1409. - PMC - PubMed
-
- Bernt KM, Armstrong SA. 2011. A role for DOT1L in MLL-rearranged leukemias. Epigenomics 3: 667–670. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
