Establishing Chromatin Regulatory Landscape during Mouse Preimplantation Development
- PMID: 27259149
- PMCID: PMC6625655
- DOI: 10.1016/j.cell.2016.05.050
Establishing Chromatin Regulatory Landscape during Mouse Preimplantation Development
Abstract
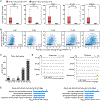
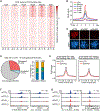
How the chromatin regulatory landscape in the inner cell mass cells is established from differentially packaged sperm and egg genomes during preimplantation development is unknown. Here, we develop a low-input DNase I sequencing (liDNase-seq) method that allows us to generate maps of DNase I-hypersensitive site (DHS) of mouse preimplantation embryos from 1-cell to morula stage. The DHS landscape is progressively established with a drastic increase at the 8-cell stage. Paternal chromatin accessibility is quickly reprogrammed after fertilization to the level similar to maternal chromatin, while imprinted genes exhibit allelic accessibility bias. We demonstrate that transcription factor Nfya contributes to zygotic genome activation and DHS formation at the 2-cell stage and that Oct4 contributes to the DHSs gained at the 8-cell stage. Our study reveals the dynamic chromatin regulatory landscape during early development and identifies key transcription factors important for DHS establishment in mammalian embryos.
Keywords: DNase I hypersensitive sites; Nfya; Oct4; chromatin accessibility; low-input DNase-seq; transcription factor; zygotic genome activation.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures





References
-
- Bell O, Tiwari VK, Thoma NH, and Schubeler D (2011). Determinants and dynamics of genome accessibility. Nature reviews Genetics 12, 554–564. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

