Direct Identification of Hundreds of Expression-Modulating Variants using a Multiplexed Reporter Assay
- PMID: 27259153
- PMCID: PMC4957403
- DOI: 10.1016/j.cell.2016.04.027
Direct Identification of Hundreds of Expression-Modulating Variants using a Multiplexed Reporter Assay
Erratum in
-
Direct Identification of Hundreds of Expression-Modulating Variants using a Multiplexed Reporter Assay.Cell. 2018 Feb 22;172(5):1132-1134. doi: 10.1016/j.cell.2018.02.021. Cell. 2018. PMID: 29474912 No abstract available.
Abstract
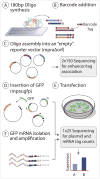
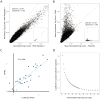
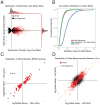
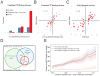
Although studies have identified hundreds of loci associated with human traits and diseases, pinpointing causal alleles remains difficult, particularly for non-coding variants. To address this challenge, we adapted the massively parallel reporter assay (MPRA) to identify variants that directly modulate gene expression. We applied it to 32,373 variants from 3,642 cis-expression quantitative trait loci and control regions. Detection by MPRA was strongly correlated with measures of regulatory function. We demonstrate MPRA's capabilities for pinpointing causal alleles, using it to identify 842 variants showing differential expression between alleles, including 53 well-annotated variants associated with diseases and traits. We investigated one in detail, a risk allele for ankylosing spondylitis, and provide direct evidence of a non-coding variant that alters expression of the prostaglandin EP4 receptor. These results create a resource of concrete leads and illustrate the promise of this approach for comprehensively interrogating how non-coding polymorphism shapes human biology.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


References
-
- Arnold, Gerlach, Stelzer, Boryn, Rath, Stark Genome-Wide Quantitative Enhancer Activity Maps Identified by STARR-seq. Science. 2013;339:1074–1077. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

