Greater Than the Sum of Parts: Complexity of the Dynamic Epigenome
- PMID: 27259201
- PMCID: PMC4898265
- DOI: 10.1016/j.molcel.2016.05.004
Greater Than the Sum of Parts: Complexity of the Dynamic Epigenome
Erratum in
-
Greater Than the Sum of Parts: Complexity of the Dynamic Epigenome.Mol Cell. 2018 Feb 1;69(3):533. doi: 10.1016/j.molcel.2018.01.015. Mol Cell. 2018. PMID: 29395069 Free PMC article. No abstract available.
Abstract
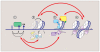
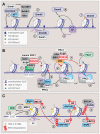
Information encoded in DNA is interpreted, modified, and propagated as chromatin. The diversity of inputs encountered by eukaryotic genomes demands a matching capacity for transcriptional outcomes provided by the combinatorial and dynamic nature of epigenetic processes. Advances in genome editing, visualization technology, and genome-wide analyses have revealed unprecedented complexity of chromatin pathways, offering explanations to long-standing questions and presenting new challenges. Here, we review recent findings, exemplified by the emerging understanding of crossregulatory interactions within chromatin, and emphasize the pathologic outcomes of epigenetic misregulation in cancer.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


References
-
- Baubec T, Colombo DF, Wirbelauer C, Schmidt J, Burger L, Krebs AR, Akalin A, Schubeler D. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015;520:243–247. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

