Comprehensive analysis of promoter-proximal RNA polymerase II pausing across mammalian cell types
- PMID: 27259512
- PMCID: PMC4893286
- DOI: 10.1186/s13059-016-0984-2
Comprehensive analysis of promoter-proximal RNA polymerase II pausing across mammalian cell types
Abstract
Background: For many genes, RNA polymerase II stably pauses before transitioning to productive elongation. Although polymerase II pausing has been shown to be a mechanism for regulating transcriptional activation, the extent to which it is involved in control of mammalian gene expression and its relationship to chromatin structure remain poorly understood.
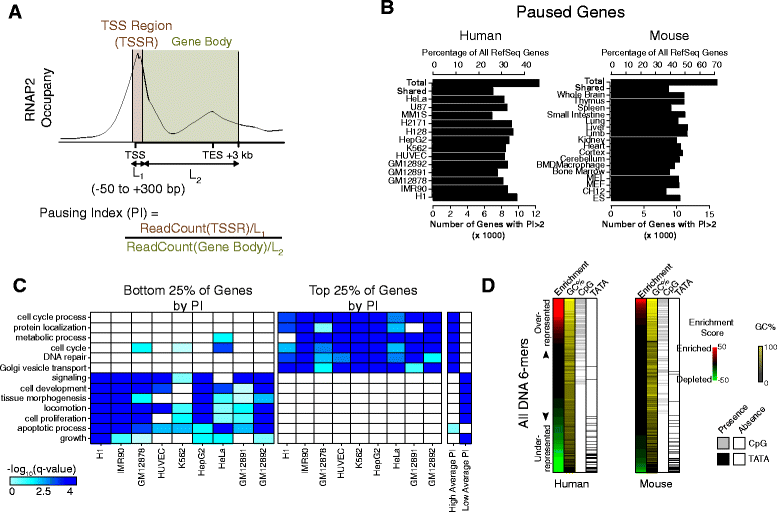
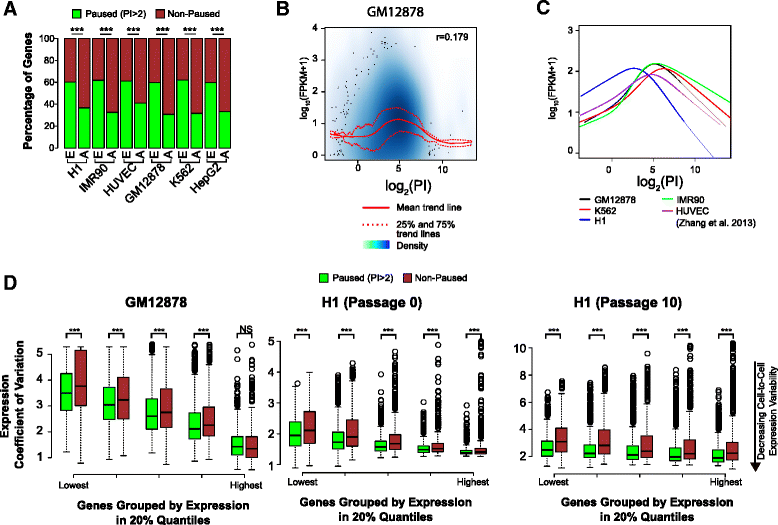
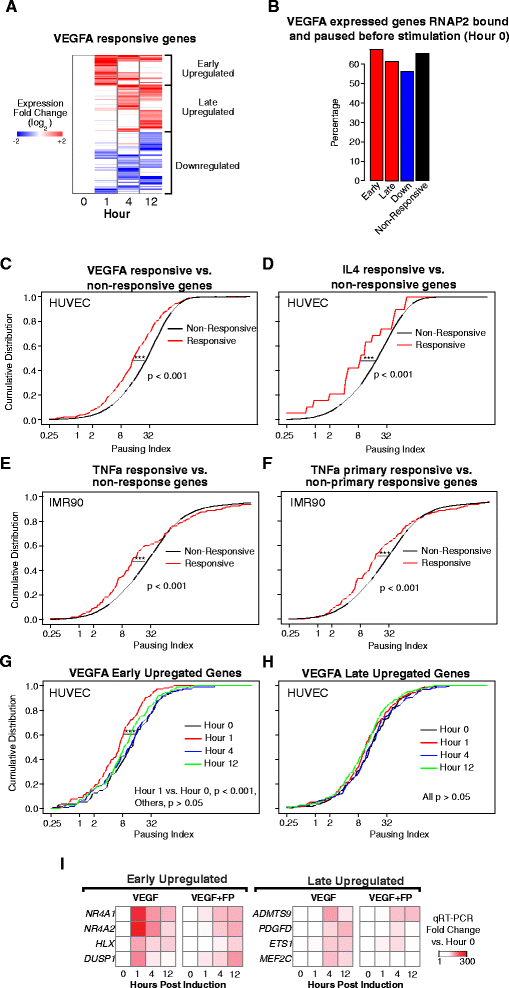
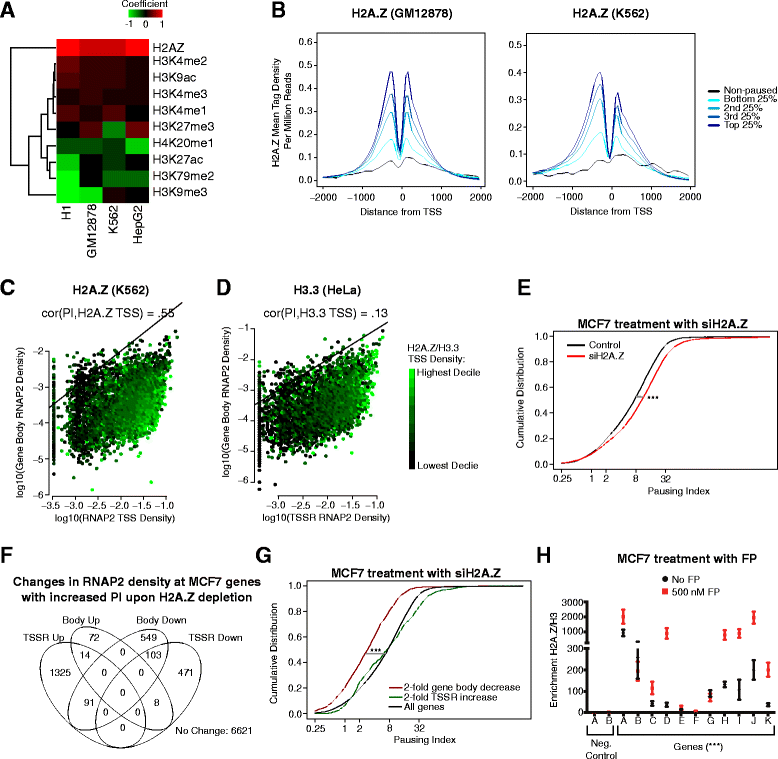
Results: Here, we analyze 85 RNA polymerase II chromatin immunoprecipitation (ChIP)-sequencing experiments from 35 different murine and human samples, as well as related genome-wide datasets, to gain new insights into the relationship between polymerase II pausing and gene regulation. Across cell and tissue types, paused genes (pausing index > 2) comprise approximately 60 % of expressed genes and are repeatedly associated with specific biological functions. Paused genes also have lower cell-to-cell expression variability. Increased pausing has a non-linear effect on gene expression levels, with moderately paused genes being expressed more highly than other paused genes. The highest gene expression levels are often achieved through a novel pause-release mechanism driven by high polymerase II initiation. In three datasets examining the impact of extracellular signals, genes responsive to stimulus have slightly lower pausing index on average than non-responsive genes, and rapid gene activation is linked to conditional pause-release. Both chromatin structure and local sequence composition near the transcription start site influence pausing, with divergent features between mammals and Drosophila. Most notably, in mammals pausing is positively correlated with histone H2A.Z occupancy at promoters.
Conclusions: Our results provide new insights into the contribution of RNA polymerase II pausing in mammalian gene regulation and chromatin structure.
Figures

Similar articles
-
Stably paused genes revealed through inhibition of transcription initiation by the TFIIH inhibitor triptolide.Genes Dev. 2015 Jan 1;29(1):39-47. doi: 10.1101/gad.246173.114. Genes Dev. 2015. PMID: 25561494 Free PMC article.
-
CBP Regulates Recruitment and Release of Promoter-Proximal RNA Polymerase II.Mol Cell. 2017 Nov 2;68(3):491-503.e5. doi: 10.1016/j.molcel.2017.09.031. Epub 2017 Oct 19. Mol Cell. 2017. PMID: 29056321 Free PMC article.
-
Origin of RNA Polymerase II pause in eumetazoans: Insights from Hydra.J Biosci. 2020;45:8. doi: 10.1007/s12038-019-9979-y. J Biosci. 2020. PMID: 31965986 Free PMC article.
-
Pause & go: from the discovery of RNA polymerase pausing to its functional implications.Curr Opin Cell Biol. 2017 Jun;46:72-80. doi: 10.1016/j.ceb.2017.03.002. Epub 2017 Mar 28. Curr Opin Cell Biol. 2017. PMID: 28363125 Free PMC article. Review.
-
RNA polymerase II pausing as a context-dependent reader of the genome.Biochem Cell Biol. 2016 Feb;94(1):82-92. doi: 10.1139/bcb-2015-0045. Epub 2015 Sep 15. Biochem Cell Biol. 2016. PMID: 26555214 Free PMC article. Review.
Cited by
-
Topoisomerase IIα represses transcription by enforcing promoter-proximal pausing.Cell Rep. 2021 Apr 13;35(2):108977. doi: 10.1016/j.celrep.2021.108977. Cell Rep. 2021. PMID: 33852840 Free PMC article.
-
Dynamic RNA Polymerase II Recruitment Drives Differentiation of the Intestine under the direction of HNF4.bioRxiv [Preprint]. 2023 Nov 10:2023.11.08.566322. doi: 10.1101/2023.11.08.566322. bioRxiv. 2023. Update in: Cell Rep. 2024 Jun 25;43(6):114242. doi: 10.1016/j.celrep.2024.114242. PMID: 37986803 Free PMC article. Updated. Preprint.
-
DSIF modulates RNA polymerase II occupancy according to template G + C content.NAR Genom Bioinform. 2022 Jul 27;4(3):lqac054. doi: 10.1093/nargab/lqac054. eCollection 2022 Sep. NAR Genom Bioinform. 2022. PMID: 35910045 Free PMC article.
-
VEGF amplifies transcription through ETS1 acetylation to enable angiogenesis.Nat Commun. 2017 Aug 29;8(1):383. doi: 10.1038/s41467-017-00405-x. Nat Commun. 2017. PMID: 28851877 Free PMC article.
-
SPT6 promotes epidermal differentiation and blockade of an intestinal-like phenotype through control of transcriptional elongation.Nat Commun. 2021 Feb 4;12(1):784. doi: 10.1038/s41467-021-21067-w. Nat Commun. 2021. PMID: 33542242 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

