16S rRNA amplicon sequencing identifies microbiota associated with oral cancer, human papilloma virus infection and surgical treatment
- PMID: 27259999
- PMCID: PMC5239478
- DOI: 10.18632/oncotarget.9710
16S rRNA amplicon sequencing identifies microbiota associated with oral cancer, human papilloma virus infection and surgical treatment
Abstract
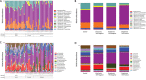
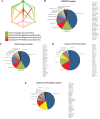
Systemic inflammatory events and localized disease, mediated by the microbiome, may be measured in saliva as head and neck squamous cell carcinoma (HNSCC) diagnostic and prognostic biomonitors. We used a 16S rRNA V3-V5 marker gene approach to compare the saliva microbiome in DNA isolated from Oropharyngeal (OPSCC), Oral Cavity Squamous Cell Carcinoma (OCSCC) patients and normal epithelium controls, to characterize the HNSCC saliva microbiota and examine their abundance before and after surgical resection.The analyses identified a predominance of Firmicutes, Proteobacteria and Bacteroidetes, with less frequent presence of Actinobacteria and Fusobacteria before surgery. At lower taxonomic levels, the most abundant genera were Streptococcus, Prevotella, Haemophilus, Lactobacillus and Veillonella, with lower numbers of Citrobacter and Neisseraceae genus Kingella. HNSCC patients had a significant loss in richness and diversity of microbiota species (p<0.05) compared to the controls. Overall, the Operational Taxonomic Units network shows that the relative abundance of OTU's within genus Streptococcus, Dialister, and Veillonella can be used to discriminate tumor from control samples (p<0.05). Tumor samples lost Neisseria, Aggregatibacter (Proteobacteria), Haemophillus (Firmicutes) and Leptotrichia (Fusobacteria). Paired taxa within family Enterobacteriaceae, together with genus Oribacterium, distinguish OCSCC samples from OPSCC and normal samples (p<0.05). Similarly, only HPV positive samples have an abundance of genus Gemellaceae and Leuconostoc (p<0.05). Longitudinal analyses of samples taken before and after surgery, revealed a reduction in the alpha diversity measure after surgery, together with an increase of this measure in patients that recurred (p<0.05). These results suggest that microbiota may be used as HNSCC diagnostic and prognostic biomonitors.
Keywords: 16s rRNA; human papilloma virus (HPV); microbiome; oral cancer; oropharyngeal cancer.
Conflict of interest statement
The authors have no conflicts of interest with the subject matter or materials discussed in this manuscript.
Figures





References
-
- Preza D, Thiede B, Olsen I, Grinde B. The proteome of the human parotid gland secretion in elderly with and without root caries. Acta odontologica Scandinavica. 2009;67:161–169. - PubMed
-
- Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, Socransky SS, Hasturk H, Van Dyke TE, Dewhirst F, Paster BJ. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. Journal of periodontology. 2009;80:1421–1432. - PMC - PubMed
-
- Fujinaka H, Takeshita T, Sato H, Yamamoto T, Nakamura J, Hase T, Yamashita Y. Relationship of periodontal clinical parameters with bacterial composition in human dental plaque. Archives of microbiology. 2013;195:371–383. - PubMed
-
- Abiko Y, Sato T, Mayanagi G, Takahashi N. Profiling of subgingival plaque biofilm microflora from periodontally healthy subjects and from subjects with periodontitis using quantitative real-time PCR. Journal of periodontal research. 2010;45:389–395. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

