A Modular, Tn7-Based System for Making Bioluminescent or Fluorescent Salmonella and Escherichia coli Strains
- PMID: 27260360
- PMCID: PMC4968541
- DOI: 10.1128/AEM.01346-16
A Modular, Tn7-Based System for Making Bioluminescent or Fluorescent Salmonella and Escherichia coli Strains
Abstract
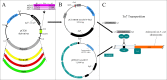
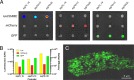
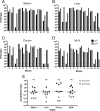
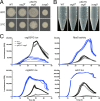
Our goal was to develop a robust tagging method that can be used to track bacterial strains in vivo To address this challenge, we adapted two existing systems: a modular plasmid-based reporter system (pCS26) that has been used for high-throughput gene expression studies in Salmonella and Escherichia coli and Tn7 transposition. We generated kanamycin- and chloramphenicol-resistant versions of pCS26 with bacterial luciferase, green fluorescent protein (GFP), and mCherry reporters under the control of σ(70)-dependent promoters to provide three different levels of constitutive expression. We improved upon the existing Tn7 system by modifying the delivery vector to accept pCS26 constructs and moving the transposase genes from a nonreplicating helper plasmid into a temperature-sensitive plasmid that can be conditionally maintained. This resulted in a 10- to 30-fold boost in transposase gene expression and transposition efficiencies of 10(-8) to 10(-10) in Salmonella enterica serovar Typhimurium and E. coli APEC O1, whereas the existing Tn7 system yielded no successful transposition events. The new reporter strains displayed reproducible signaling in microwell plate assays, confocal microscopy, and in vivo animal infections. We have combined two flexible and complementary tools that can be used for a multitude of molecular biology applications within the Enterobacteriaceae This system can accommodate new promoter-reporter combinations as they become available and can help to bridge the gap between modern, high-throughput technologies and classical molecular genetics.
Importance: This article describes a flexible and efficient system for tagging bacterial strains. Using our modular plasmid system, a researcher can easily change the reporter type or the promoter driving expression and test the parameters of these new constructs in vitro Selected constructs can then be stably integrated into the chromosomes of desired strains in two simple steps. We demonstrate the use of this system in Salmonella and E. coli, and we predict that it will be widely applicable to other bacterial strains within the Enterobacteriaceae This technology will allow for improved in vivo analysis of bacterial pathogens.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Tn5/7-lux: a versatile tool for the identification and capture of promoters in gram-negative bacteria.BMC Microbiol. 2015 Feb 4;15(1):17. doi: 10.1186/s12866-015-0354-3. BMC Microbiol. 2015. PMID: 25648327 Free PMC article.
-
An improved Tn7-lux reporter for broad host range, chromosomally-integrated promoter fusions in Gram-negative bacteria.J Microbiol Methods. 2015 Nov;118:75-7. doi: 10.1016/j.mimet.2015.08.016. Epub 2015 Sep 1. J Microbiol Methods. 2015. PMID: 26341612 Free PMC article.
-
A simple method for construction of pir+ Enterobacterial hosts for maintenance of R6K replicon plasmids.BMC Res Notes. 2012 Mar 20;5:157. doi: 10.1186/1756-0500-5-157. BMC Res Notes. 2012. PMID: 22433797 Free PMC article.
-
New methods in Salmonella genetics.Microbiologia. 1994 Dec;10(4):357-70. Microbiologia. 1994. PMID: 7772291 Review.
-
Molecular Profiling: Catecholamine Modulation of Gene Expression in Escherichia coli O157:H7 and Salmonella enterica Serovar Typhimurium.Adv Exp Med Biol. 2016;874:167-82. doi: 10.1007/978-3-319-20215-0_7. Adv Exp Med Biol. 2016. PMID: 26589218 Review.
Cited by
-
Parallel evolution leading to impaired biofilm formation in invasive Salmonella strains.PLoS Genet. 2019 Jun 24;15(6):e1008233. doi: 10.1371/journal.pgen.1008233. eCollection 2019 Jun. PLoS Genet. 2019. PMID: 31233504 Free PMC article.
-
Characterization of colonization kinetics and virulence potential of Salmonella Enteritidis in chickens by photonic detection.Front Vet Sci. 2022 Aug 2;9:948448. doi: 10.3389/fvets.2022.948448. eCollection 2022. Front Vet Sci. 2022. PMID: 35982923 Free PMC article.
-
Reconstructing promoter activity from Lux bioluminescent reporters.PLoS Comput Biol. 2017 Sep 18;13(9):e1005731. doi: 10.1371/journal.pcbi.1005731. eCollection 2017 Sep. PLoS Comput Biol. 2017. PMID: 28922354 Free PMC article.
-
Flagellar-based motility accelerates IgA-mediated agglutination of Salmonella Typhimurium at high bacterial cell densities.Front Immunol. 2023 May 18;14:1193855. doi: 10.3389/fimmu.2023.1193855. eCollection 2023. Front Immunol. 2023. PMID: 37275888 Free PMC article.
-
A SNP in the Cache 1 Signaling Domain of Diguanylate Cyclase STM1987 Leads to Increased In Vivo Fitness of Invasive Salmonella Strains.Infect Immun. 2021 Mar 17;89(4):e00810-20. doi: 10.1128/IAI.00810-20. Print 2021 Mar 17. Infect Immun. 2021. PMID: 33468583 Free PMC article.
References
-
- Porwollik S, Santiviago CA, Cheng P, Long F, Desai P, Fredlund J, Srikumar S, Silva CA, Chu W, Chen X, Canals R, Reynolds MM, Bogomolnaya L, Shields C, Cui P, Guo J, Zheng Y, Endicott-Yazdani T, Yang HJ, Maple A, Ragoza Y, Blondel CJ, Valenzuela C, Andrews-Polymenis H, McClelland M. 2014. Defined single-gene and multi-gene deletion mutant collections in Salmonella enterica sv Typhimurium. PLoS One 9:e99820. doi:10.1371/journal.pone.0099820. - DOI - PMC - PubMed
-
- Santiviago CA, Reynolds MM, Porwollik S, Choi SH, Long F, Andrews-Polymenis HL, McClelland M. 2009. Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog 5:e1000477. doi:10.1371/journal.ppat.1000477. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

