Computational modeling of Repeat1 region of INI1/hSNF5: An evolutionary link with ubiquitin
- PMID: 27261671
- PMCID: PMC5338235
- DOI: 10.1002/pro.2961
Computational modeling of Repeat1 region of INI1/hSNF5: An evolutionary link with ubiquitin
Abstract
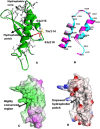
The structure of a protein can be very informative of its function. However, determining protein structures experimentally can often be very challenging. Computational methods have been used successfully in modeling structures with sufficient accuracy. Here we have used computational tools to predict the structure of an evolutionarily conserved and functionally significant domain of Integrase interactor (INI)1/hSNF5 protein. INI1 is a component of the chromatin remodeling SWI/SNF complex, a tumor suppressor and is involved in many protein-protein interactions. It belongs to SNF5 family of proteins that contain two conserved repeat (Rpt) domains. Rpt1 domain of INI1 binds to HIV-1 Integrase, and acts as a dominant negative mutant to inhibit viral replication. Rpt1 domain also interacts with oncogene c-MYC and modulates its transcriptional activity. We carried out an ab initio modeling of a segment of INI1 protein containing the Rpt1 domain. The structural model suggested the presence of a compact and well defined ββαα topology as core structure in the Rpt1 domain of INI1. This topology in Rpt1 was similar to PFU domain of Phospholipase A2 Activating Protein, PLAA. Interestingly, PFU domain shares similarity with Ubiquitin and has ubiquitin binding activity. Because of the structural similarity between Rpt1 domain of INI1 and PFU domain of PLAA, we propose that Rpt1 domain of INI1 may participate in ubiquitin recognition or binding with ubiquitin or ubiquitin related proteins. This modeling study may shed light on the mode of interactions of Rpt1 domain of INI1 and is likely to facilitate future functional studies of INI1.
Keywords: SNF5; integrase interactor (INI)1/hSNF5; protein structure modeling; repeats; ubiquitin.
© 2016 The Protein Society.
Figures

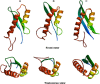


Similar articles
-
INI1/SMARCB1 Rpt1 domain mimics TAR RNA in binding to integrase to facilitate HIV-1 replication.Nat Commun. 2021 May 12;12(1):2743. doi: 10.1038/s41467-021-22733-9. Nat Commun. 2021. PMID: 33980829 Free PMC article.
-
The structure of INI1/hSNF5 RPT1 and its interactions with the c-MYC:MAX heterodimer provide insights into the interplay between MYC and the SWI/SNF chromatin remodeling complex.FEBS J. 2018 Nov;285(22):4165-4180. doi: 10.1111/febs.14660. Epub 2018 Oct 1. FEBS J. 2018. PMID: 30222246 Free PMC article.
-
c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function.Nat Genet. 1999 May;22(1):102-5. doi: 10.1038/8811. Nat Genet. 1999. PMID: 10319872
-
Integrase interactor 1 in health and disease.Curr Protein Pept Sci. 2015;16(6):478-90. doi: 10.2174/1389203716666150316115156. Curr Protein Pept Sci. 2015. PMID: 25772157 Review.
-
The role of INI1/hSNF5 in gene regulation and cancer.Biochem Cell Biol. 2009 Feb;87(1):163-77. doi: 10.1139/O08-113. Biochem Cell Biol. 2009. PMID: 19234532 Review.
Cited by
-
SMARCB1 Promotes Ubiquitination and Degradation of NR4A3 via Direct Interaction Driven by ROS in Vascular Endothelial Cell Injury.Oxid Med Cell Longev. 2020 Oct 23;2020:2048210. doi: 10.1155/2020/2048210. eCollection 2020. Oxid Med Cell Longev. 2020. PMID: 33163142 Free PMC article.
-
Human chromatin remodelers regulating HIV-1 transcription: a target for small molecule inhibitors.Epigenetics Chromatin. 2025 Apr 16;18(1):21. doi: 10.1186/s13072-025-00582-w. Epigenetics Chromatin. 2025. PMID: 40241225 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

