Two Mutually Exclusive Local Chromatin States Drive Efficient V(D)J Recombination
- PMID: 27264181
- PMCID: PMC4914699
- DOI: 10.1016/j.celrep.2016.05.020
Two Mutually Exclusive Local Chromatin States Drive Efficient V(D)J Recombination
Abstract
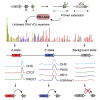
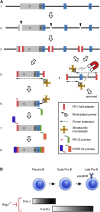
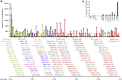
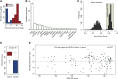
Variable (V), diversity (D), and joining (J) (V(D)J) recombination is the first determinant of antigen receptor diversity. Understanding how recombination is regulated requires a comprehensive, unbiased readout of V gene usage. We have developed VDJ sequencing (VDJ-seq), a DNA-based next-generation-sequencing technique that quantitatively profiles recombination products. We reveal a 200-fold range of recombination efficiency among recombining V genes in the primary mouse Igh repertoire. We used machine learning to integrate these data with local chromatin profiles to identify combinatorial patterns of epigenetic features that associate with active VH gene recombination. These features localize downstream of VH genes and are excised by recombination, revealing a class of cis-regulatory element that governs recombination, distinct from expression. We detect two mutually exclusive chromatin signatures at these elements, characterized by CTCF/RAD21 and PAX5/IRF4, which segregate with the evolutionary history of associated VH genes. Thus, local chromatin signatures downstream of VH genes provide an essential layer of regulation that determines recombination efficiency.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Alamyar E., Giudicelli V., Li S., Duroux P., Lefranc M.-P. IMGT/HighV-QUEST: the IMGT web portal for immunoglobulin (IG) or antibody and T cell receptor (TR) analysis from NGS high throughput and deep sequencing. Immunome Res. 2012;8:26.
-
- Bolland D.J., Wood A.L., Johnston C.M., Bunting S.F., Morgan G., Chakalova L., Fraser P.J., Corcoran A.E. Antisense intergenic transcription in V(D)J recombination. Nat. Immunol. 2004;5:630–637. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

