Global update on the susceptibility of human influenza viruses to neuraminidase inhibitors, 2014-2015
- PMID: 27265623
- PMCID: PMC5357725
- DOI: 10.1016/j.antiviral.2016.06.001
Global update on the susceptibility of human influenza viruses to neuraminidase inhibitors, 2014-2015
Abstract
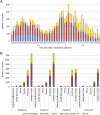
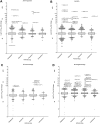
The World Health Organization (WHO) Collaborating Centres for Reference and Research on Influenza (WHO CCs) tested 13,312 viruses collected by WHO recognized National Influenza Centres between May 2014 and May 2015 to determine 50% inhibitory concentration (IC50) data for neuraminidase inhibitors (NAIs) oseltamivir, zanamivir, peramivir and laninamivir. Ninety-four per cent of the viruses tested by the WHO CCs were from three WHO regions: Western Pacific, the Americas and Europe. Approximately 0.5% (n = 68) of viruses showed either highly reduced inhibition (HRI) or reduced inhibition (RI) (n = 56) against at least one of the four NAIs. Of the twelve viruses with HRI, six were A(H1N1)pdm09 viruses, three were A(H3N2) viruses and three were B/Yamagata-lineage viruses. The overall frequency of viruses with RI or HRI by the NAIs was lower than that observed in 2013-14 (1.9%), but similar to the 2012-13 period (0.6%). Based on the current analysis, the NAIs remain an appropriate choice for the treatment and prophylaxis of influenza virus infections.
Keywords: Antiviral resistance; Global analysis; Influenza virus; Neuraminidase inhibitors; Oseltamivir; Reduced susceptibility.
Copyright © 2016 The Authors. Published by Elsevier B.V. All rights reserved.
Figures

References
-
- Abed Y., Pizzorno A., Bouhy X., Boivin G. Permissive changes in the neuraminidase play a dominant role in improving the viral fitness of oseltamivir-resistant seasonal influenza A(H1N1) strains. Antivir. Res. 2015;114:57–61. - PubMed
-
- Butler J., Hooper K.A., Petrie S., Lee R., Maurer-Stroh S., Reh L., Guarnaccia T., Baas C., Xue L., Vitesnik S., Leang S.K., McVernon J., Kelso A., Barr I.G., McCaw J.M., Bloom J.D., Hurt A.C. Estimating the fitness advantage conferred by permissive neuraminidase mutations in recent oseltamivir-resistant A(H1N1)pdm09 influenza viruses. PLoS Pathog. 2014;10:e1004065. - PMC - PubMed
-
- Deng Y.M., Spirason N., Iannello P., Jelley L., Lau H., Barr I.G. A simplified Sanger sequencing method for full genome sequencing of multiple subtypes of human influenza A viruses. J. Clin. Virol. 2015;68:43–48. - PubMed
-
- Dharan N.J., Gubareva L.V., Meyer J.J., Okomo-Adhiambo M., McClinton R.C., Marshall S.A., St G.K., Epperson S., Brammer L., Klimov A.I., Bresee J.S., Fry A.M. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States. JAMA. 2009;301:1034–1041. - PubMed
-
- Hurt A.C., Ernest J., Deng Y., Iannello P., Besselaar T.G., Birch C., Buchy P., Chittaganpitch M., Chiu S.C., Dwyer D.E., Guigon A., Harrower B., Kei I.P., Kok T., Lin C., McPhie K., Mohd A., Olveda R., Panayotou T., Rawlinson W., Scott L., Smith D., D’Souza H., Komadina N., Shaw R., Kelso A., Barr I.G. Emergence and spread of oseltamivir-resistant A(H1N1) influenza viruses in Oceania, South East Asia and South Africa. Antivir. Res. 2009;83:90–93. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

