Structure and mechanism of assembly line polyketide synthases
- PMID: 27266330
- PMCID: PMC5136517
- DOI: 10.1016/j.sbi.2016.05.009
Structure and mechanism of assembly line polyketide synthases
Abstract
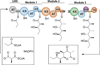
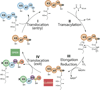
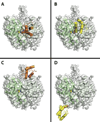
Assembly line polyketide synthases (PKSs) are remarkable biosynthetic machines with considerable potential for structure-based engineering. Several types of protein-protein interactions, both within and between PKS modules, play important roles in the catalytic cycle of a multimodular PKS. Additionally, vectorial biosynthesis is enabled by the energetic coupling of polyketide chain elongation to the channeling of intermediates between successive modules. A combination of high-resolution analysis of smaller PKS components and lower resolution characterization of intact modules and bimodules has yielded insights into the structure and organization of a prototypical assembly line PKS. This review discusses our understanding of key structure-function relationships in this family of megasynthases, along with a recap of key unanswered questions in the field.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures
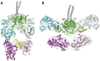
References
-
- Walsh CT. Polyketide and Nonribosomal Peptide Antibiotics: Modularity and Versatility. Science. 2004;303:1805–1810. - PubMed
-
- Hopwood DA. Streptomyces in Nature and Medicine. 2007:1–261.
-
- Khosla C, Tang Y, Chen AY, Schnarr NA, Cane DE. Structure and Mechanism of the 6-Deoxyerythronolide B Synthase. Annu. Rev. Biochem. 2007;76:195–221. - PubMed
-
- Caffrey P, Bevitt DJ, Staunton J, Leadlay PF. Identification of DEBS 1, DEBS 2 and DEBS 3, the multienzyme polypeptides of the erythromycin-producing polyketide synthase from Saccharopolyspora erythraea. FEBS Journal. 1992;304:225–228. - PubMed
-
- Gokhale RS, Lau J, Cane DE, Khosla C. Functional Orientation of the Acyltransferase Domain in a Module of the Erythromycin Polyketide Synthase. Biochemistry. 1998;37:2524–2528. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

