Evaluation of rate law approximations in bottom-up kinetic models of metabolism
- PMID: 27266508
- PMCID: PMC4895898
- DOI: 10.1186/s12918-016-0283-2
Evaluation of rate law approximations in bottom-up kinetic models of metabolism
Abstract
Background: The mechanistic description of enzyme kinetics in a dynamic model of metabolism requires specifying the numerical values of a large number of kinetic parameters. The parameterization challenge is often addressed through the use of simplifying approximations to form reaction rate laws with reduced numbers of parameters. Whether such simplified models can reproduce dynamic characteristics of the full system is an important question.
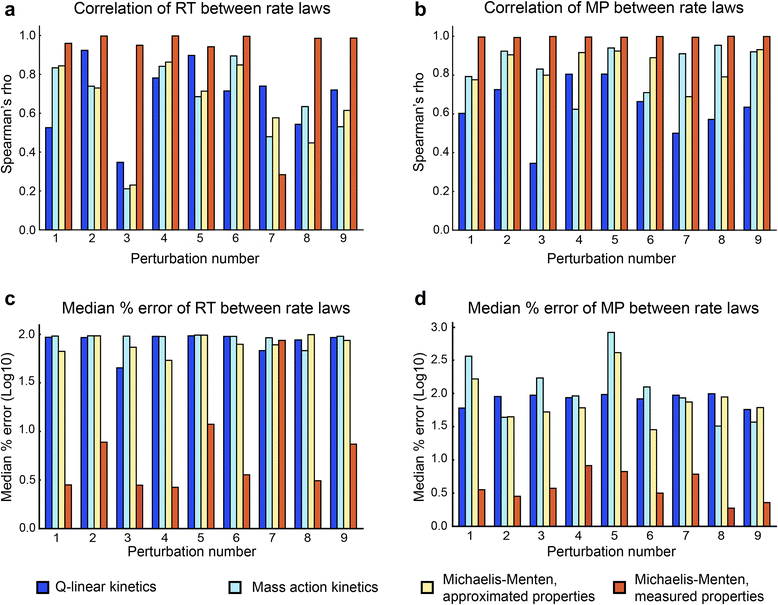
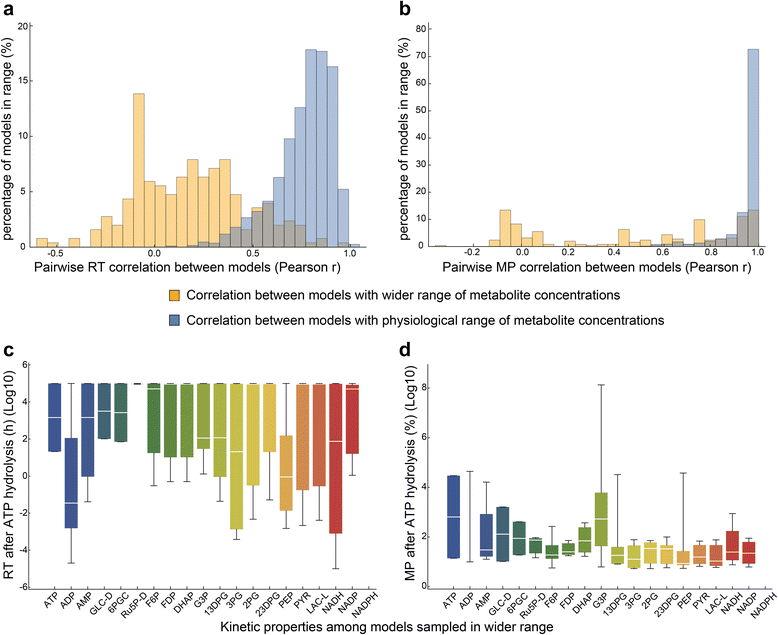
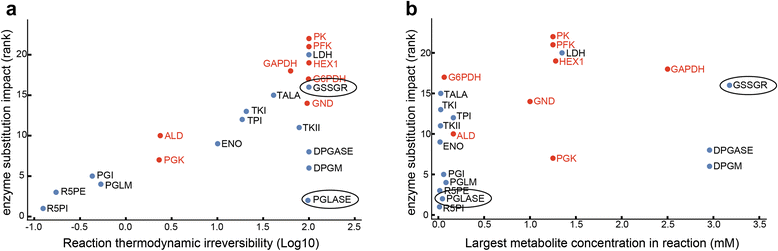
Results: In this work, we compared the local transient response properties of dynamic models constructed using rate laws with varying levels of approximation. These approximate rate laws were: 1) a Michaelis-Menten rate law with measured enzyme parameters, 2) a Michaelis-Menten rate law with approximated parameters, using the convenience kinetics convention, 3) a thermodynamic rate law resulting from a metabolite saturation assumption, and 4) a pure chemical reaction mass action rate law that removes the role of the enzyme from the reaction kinetics. We utilized in vivo data for the human red blood cell to compare the effect of rate law choices against the backdrop of physiological flux and concentration differences. We found that the Michaelis-Menten rate law with measured enzyme parameters yields an excellent approximation of the full system dynamics, while other assumptions cause greater discrepancies in system dynamic behavior. However, iteratively replacing mechanistic rate laws with approximations resulted in a model that retains a high correlation with the true model behavior. Investigating this consistency, we determined that the order of magnitude differences among fluxes and concentrations in the network were greatly influential on the network dynamics. We further identified reaction features such as thermodynamic reversibility, high substrate concentration, and lack of allosteric regulation, which make certain reactions more suitable for rate law approximations.
Conclusions: Overall, our work generally supports the use of approximate rate laws when building large scale kinetic models, due to the key role that physiologically meaningful flux and concentration ranges play in determining network dynamics. However, we also showed that detailed mechanistic models show a clear benefit in prediction accuracy when data is available. The work here should help to provide guidance to future kinetic modeling efforts on the choice of rate law and parameterization approaches.
Keywords: Approximate rate laws; Kinetic modeling; Mass action kinetics; Metabolic modeling; Michaelis-Menten kinetics.
Figures



Similar articles
-
Hybrid dynamic modeling of Escherichia coli central metabolic network combining Michaelis-Menten and approximate kinetic equations.Biosystems. 2010 May;100(2):150-7. doi: 10.1016/j.biosystems.2010.03.001. Epub 2010 Mar 10. Biosystems. 2010. PMID: 20226228
-
Bringing metabolic networks to life: convenience rate law and thermodynamic constraints.Theor Biol Med Model. 2006 Dec 15;3:41. doi: 10.1186/1742-4682-3-41. Theor Biol Med Model. 2006. PMID: 17173669 Free PMC article.
-
Modular rate laws for enzymatic reactions: thermodynamics, elasticities and implementation.Bioinformatics. 2010 Jun 15;26(12):1528-34. doi: 10.1093/bioinformatics/btq141. Epub 2010 Apr 12. Bioinformatics. 2010. PMID: 20385728
-
New types of experimental data shape the use of enzyme kinetics for dynamic network modeling.FEBS J. 2014 Jan;281(2):549-71. doi: 10.1111/febs.12525. Epub 2013 Nov 4. FEBS J. 2014. PMID: 24034816 Review.
-
Validity of the Michaelis-Menten equation--steady-state or reactant stationary assumption: that is the question.FEBS J. 2014 Jan;281(2):464-72. doi: 10.1111/febs.12564. Epub 2013 Nov 18. FEBS J. 2014. PMID: 24245583 Review.
Cited by
-
Network-level allosteric effects are elucidated by detailing how ligand-binding events modulate utilization of catalytic potentials.PLoS Comput Biol. 2018 Aug 7;14(8):e1006356. doi: 10.1371/journal.pcbi.1006356. eCollection 2018 Aug. PLoS Comput Biol. 2018. PMID: 30086174 Free PMC article.
-
Modular assembly of dynamic models in systems biology.PLoS Comput Biol. 2021 Oct 13;17(10):e1009513. doi: 10.1371/journal.pcbi.1009513. eCollection 2021 Oct. PLoS Comput Biol. 2021. PMID: 34644304 Free PMC article.
-
Approaches to Computational Strain Design in the Multiomics Era.Front Microbiol. 2019 Apr 5;10:597. doi: 10.3389/fmicb.2019.00597. eCollection 2019. Front Microbiol. 2019. PMID: 31024467 Free PMC article.
-
Metabolomics and modelling approaches for systems metabolic engineering.Metab Eng Commun. 2022 Oct 15;15:e00209. doi: 10.1016/j.mec.2022.e00209. eCollection 2022 Dec. Metab Eng Commun. 2022. PMID: 36281261 Free PMC article. Review.
-
Metabolic kinetic modeling provides insight into complex biological questions, but hurdles remain.Curr Opin Biotechnol. 2019 Oct;59:24-30. doi: 10.1016/j.copbio.2019.02.005. Epub 2019 Mar 7. Curr Opin Biotechnol. 2019. PMID: 30851632 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

