Gigwa-Genotype investigator for genome-wide analyses
- PMID: 27267926
- PMCID: PMC4897896
- DOI: 10.1186/s13742-016-0131-8
Gigwa-Genotype investigator for genome-wide analyses
Erratum in
-
Erratum to: Gigwa-Genotype investigator for genome-wide analyses.Gigascience. 2016 Nov 2;5(1):48. doi: 10.1186/s13742-016-0153-2. Gigascience. 2016. PMID: 27806726 Free PMC article. No abstract available.
Abstract
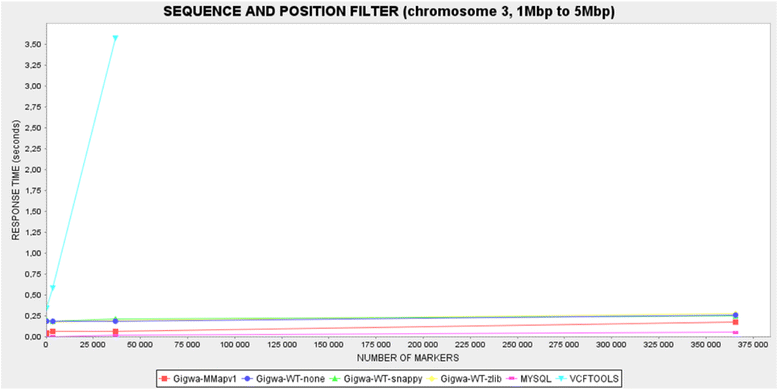
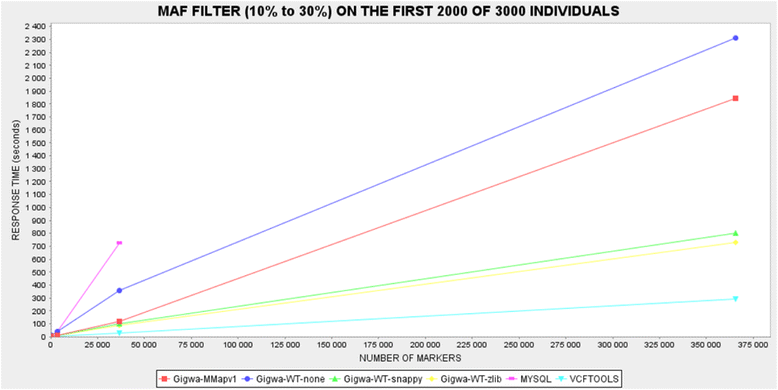
Background: Exploring the structure of genomes and analyzing their evolution is essential to understanding the ecological adaptation of organisms. However, with the large amounts of data being produced by next-generation sequencing, computational challenges arise in terms of storage, search, sharing, analysis and visualization. This is particularly true with regards to studies of genomic variation, which are currently lacking scalable and user-friendly data exploration solutions.
Description: Here we present Gigwa, a web-based tool that provides an easy and intuitive way to explore large amounts of genotyping data by filtering it not only on the basis of variant features, including functional annotations, but also on genotype patterns. The data storage relies on MongoDB, which offers good scalability properties. Gigwa can handle multiple databases and may be deployed in either single- or multi-user mode. In addition, it provides a wide range of popular export formats.
Conclusions: The Gigwa application is suitable for managing large amounts of genomic variation data. Its user-friendly web interface makes such processing widely accessible. It can either be simply deployed on a workstation or be used to provide a shared data portal for a given community of researchers.
Keywords: Genomic variations; HapMap; INDEL; MongoDB; NoSQL; SNP; VCF; Web interface.
Figures


References
-
- The 3000 rice genomes project. The 3,000 rice genomes project. Gigascience. 2014; 3:7. http://dx.doi.org/10.1186/2047-217X-3-7
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

