Process optimization for production and purification of a thermostable, organic solvent tolerant lipase from Acinetobacter sp. AU07
- PMID: 27268114
- PMCID: PMC4927683
- DOI: 10.1016/j.bjm.2015.04.002
Process optimization for production and purification of a thermostable, organic solvent tolerant lipase from Acinetobacter sp. AU07
Abstract
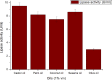
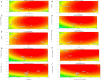
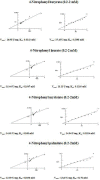
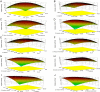
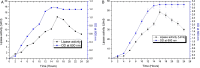
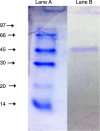
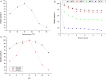
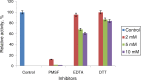
The purpose of this study was to isolate, purify and optimize the production conditions of an organic solvent tolerant and thermostable lipase from Acinetobacter sp. AU07 isolated from distillery waste. The lipase production was optimized by response surface methodology, and a maximum production of 14.5U/mL was observed at 30°C and pH 7, using a 0.5% (v/v) inoculum, 2% (v/v) castor oil (inducer), and agitation 150rpm. The optimized conditions from the shake flask experiments were validated in a 3L lab scale bioreactor, and the lipase production increased to 48U/mL. The enzyme was purified by ammonium sulfate precipitation and ion exchange chromatography and the overall yield was 36%. SDS-PAGE indicated a molecular weight of 45kDa for the purified protein, and Matrix assisted laser desorption/ionization time of flight analysis of the purified lipase showed sequence similarity with GDSL family of lipases. The optimum temperature and pH for activity of the enzyme was found to be 50°C and 8.0, respectively. The lipase was completely inhibited by phenylmethylsulfonyl fluoride but minimal inhibition was observed when incubated with ethylenediaminetetraacetic acid and dithiothreitol. The enzyme was stable in the presence of non-polar hydrophobic solvents. Detergents like SDS inhibited enzyme activity; however, there was minimal loss of enzyme activity when incubated with hydrogen peroxide, Tween 80 and Triton X-100. The kinetic constants (Km and Vmax) revealed that the hydrolytic activity of the lipase was specific to moderate chain fatty acid esters. The Vmax, Km and Vmax/Km ratio of the enzyme were 16.98U/mg, 0.51mM, and 33.29, respectively when 4-nitrophenyl palmitate was used as a substrate.
Keywords: Acinetobacter sp.; MALDI-TOF; Organic solvent tolerant lipase; Response surface methodology; Thermostable lipase.
Copyright © 2016 Sociedade Brasileira de Microbiologia. Published by Elsevier Editora Ltda. All rights reserved.
Figures




Similar articles
-
Isolation and characterization of a novel thermophilic-organic solvent stable lipase from Acinetobacter baylyi.Appl Biochem Biotechnol. 2010 Nov;162(5):1362-76. doi: 10.1007/s12010-010-8928-x. Epub 2010 Feb 24. Appl Biochem Biotechnol. 2010. PMID: 20177822
-
Purification of lipase from Aspergillus fumigatus using Octyl Sepharose column chromatography and its characterization.J Basic Microbiol. 2018 Oct;58(10):857-866. doi: 10.1002/jobm.201800129. Epub 2018 Jul 24. J Basic Microbiol. 2018. PMID: 30039877
-
A new thermostable and organic solvent-tolerant lipase from Staphylococcus warneri; optimization of media and production conditions using statistical methods.Appl Biochem Biotechnol. 2015 Jan;175(2):855-69. doi: 10.1007/s12010-014-1331-2. Epub 2014 Oct 25. Appl Biochem Biotechnol. 2015. PMID: 25344436
-
Organic solvent tolerant lipases and applications.ScientificWorldJournal. 2014 Feb 2;2014:625258. doi: 10.1155/2014/625258. eCollection 2014. ScientificWorldJournal. 2014. PMID: 24672342 Free PMC article. Review.
-
Synthesis of esters by immobilized-lipase-catalyzed condensation reaction of sugars and fatty acids in water-miscible organic solvent.J Biosci Bioeng. 2005 Feb;99(2):87-94. doi: 10.1263/jbb.99.87. J Biosci Bioeng. 2005. PMID: 16233762 Review.
Cited by
-
Evaluation of the Impact of Esterases and Lipases from the Circulatory System against Substrates of Different Lipophilicity.Int J Mol Sci. 2022 Jan 23;23(3):1262. doi: 10.3390/ijms23031262. Int J Mol Sci. 2022. PMID: 35163184 Free PMC article.
-
Optimization of organic solvent-tolerant lipase production by Acinetobacter sp. UBT1 using deoiled castor seed cake.3 Biotech. 2020 Dec;10(12):508. doi: 10.1007/s13205-020-02501-0. Epub 2020 Nov 5. 3 Biotech. 2020. PMID: 33184594 Free PMC article.
-
Characterization of a Stable Form of Carboxypeptidase G2 (Glucarpidase), a Potential Biobetter Variant, From Acinetobacter sp. 263903-1.Mol Biotechnol. 2021 Dec;63(12):1155-1168. doi: 10.1007/s12033-021-00370-3. Epub 2021 Jul 15. Mol Biotechnol. 2021. PMID: 34268672
-
Realm of Thermoalkaline Lipases in Bioprocess Commodities.J Lipids. 2018 Feb 14;2018:5659683. doi: 10.1155/2018/5659683. eCollection 2018. J Lipids. 2018. PMID: 29666707 Free PMC article. Review.
-
Lipase from Rhizopus oryzae R1: in-depth characterization, immobilization, and evaluation in biodiesel production.J Genet Eng Biotechnol. 2021 Jan 5;19(1):1. doi: 10.1186/s43141-020-00094-y. J Genet Eng Biotechnol. 2021. PMID: 33400043 Free PMC article.
References
-
- Maruyama T., Nakajima M., Ichikawa S., Nabetani H., Furusaki S., Seki M. Oil–water interfacial activation of lipase for interesterification of triglyceride and fatty acid. J Am Oil Chem Soc. 2000;77:1121–1127.
-
- Verma M.L., Azmi W., Kanwar S.S. Microbial lipases: at the interface of aqueous and non-aqueous media. A review. Acta Microbiol Immunol Hung. 2008;55:265–294. - PubMed
-
- Belfrage P., Fredrikson G., Stralfors P., Tornquist H. In: Lipases. Borgstrom B., Brockman H.L., editors. Elsevier Science; Amsterdam: 1984. pp. 365–416.
-
- Akoh C.C., Sellappan S., Fomuso L.B., Yankah G V.V. Enzymatic synthesis of structured lipids. In: Kuo T.M., Gardner H.W., editors. Lipid Biotechnology. Marcel Dekker; New York, USA: 2002. pp. 433–460.
-
- Saxena R.K., Sheoran A., Giri B., Davidson S.W. Purification strategies for microbial lipases. J Microbiol Methods. 2003;52:1–18. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
