Dispensable chromosomes in Fusarium oxysporum f. sp. lycopersici
- PMID: 27271322
- PMCID: PMC6638487
- DOI: 10.1111/mpp.12440
Dispensable chromosomes in Fusarium oxysporum f. sp. lycopersici
Abstract
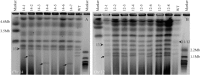
The genomes of many filamentous fungi consist of a 'core' part containing conserved genes essential for normal development as well as conditionally dispensable (CD) or lineage-specific (LS) chromosomes. In the plant-pathogenic fungus Fusarium oxysporum f. sp. lycopersici, one LS chromosome harbours effector genes that contribute to pathogenicity. We employed flow cytometry to select for events of spontaneous (partial) loss of either the two smallest LS chromosomes or two different core chromosomes. We determined the rate of spontaneous loss of the 'effector' LS chromosome in vitro at around 1 in 35 000 spores. In addition, a viable strain was obtained lacking chromosome 12, which is considered to be a part of the core genome. We also isolated strains carrying approximately 1-Mb deletions in the LS chromosomes and in the dispensable core chromosome. The large core chromosome 1 was never observed to sustain deletions over 200 kb. Whole-genome sequencing revealed that some of the sites at which the deletions occurred were the same in several independent strains obtained for the two chromosomes tested, indicating the existence of deletion hotspots. For the core chromosome, this deletion hotspot was the site of insertion of the marker used to select for loss events. Loss of the core chromosome did not affect pathogenicity, whereas loss of the effector chromosome led to a complete loss of pathogenicity.
Keywords: chromosome deletions; conditionally dispensable chromosomes; flow cytometry; pathogenic fungi.
© 2016 THE AUTHORS. MOLECULAR PLANT PATHOLOGY PUBLISHED BY BRITISH SOCIETY FOR PLANT PATHOLOGY AND JOHN WILEY & SONS LTD.
Figures




References
-
- Balesdent, M. , Fudal, I. , Ollivier, B. , Bally, P. , Grandaubert, J. , Eber, F. , Chèvre, A. , Leflon, M. and Rouxel, T. (2013) The dispensable chromosome of Leptosphaeria maculans shelters an effector gene conferring avirulence towards Brassica rapa . New Phytol. 198, 887–898. - PubMed
-
- Chuma, I. , Isobe, C. , Hotta, Y. , Ibaragi, K. , Futamata, N. , Kusaba, M. , Yoshida, K. , Terauchi, R. , Fujita, Y. and Nakayashiki, H. (2011) Multiple translocation of the AVR‐pita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and related species. PLoS Pathog. 7, e1002147. - PMC - PubMed
-
- Covert, S.F. (1998) Supernumerary chromosomes in filamentous fungi. Curr. Genet. 33, 311–319. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

