The Role of microRNAs in the Pathogenesis of Herpesvirus Infection
- PMID: 27271654
- PMCID: PMC4926176
- DOI: 10.3390/v8060156
The Role of microRNAs in the Pathogenesis of Herpesvirus Infection
Abstract
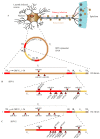
MicroRNAs (miRNAs) are small non-coding RNAs important in gene regulation. They are able to regulate mRNA translation through base-pair complementarity. Cellular miRNAs have been involved in the regulation of nearly all cellular pathways, and their deregulation has been associated with several diseases such as cancer. Given the importance of microRNAs to cell homeostasis, it is no surprise that viruses have evolved to take advantage of this cellular pathway. Viruses have been reported to be able to encode and express functional viral microRNAs that target both viral and cellular transcripts. Moreover, viral inhibition of key proteins from the microRNA pathway and important changes in cellular microRNA pool have been reported upon viral infection. In addition, viruses have developed multiple mechanisms to avoid being targeted by cellular microRNAs. This complex interaction between host and viruses to control the microRNA pathway usually favors viral infection and persistence by either reducing immune detection, avoiding apoptosis, promoting cell growth, or promoting lytic or latent infection. One of the best examples of this virus-host-microRNA interplay emanates from members of the Herperviridae family, namely the herpes simplex virus type 1 and type 2 (HSV-1 and HSV-2), human cytomegalovirus (HCMV), human herpesvirus 8 (HHV-8), and the Epstein-Barr virus (EBV). In this review, we will focus on the general functions of microRNAs and the interactions between herpesviruses, human hosts, and microRNAs and will delve into the related mechanisms that contribute to infection and pathogenesis.
Keywords: herpesvirus; immune evasion; latency; microRNAs; oncogenesis; pathogenesis.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

