Direct observation of DNA threading in flap endonuclease complexes
- PMID: 27273516
- PMCID: PMC4939078
- DOI: 10.1038/nsmb.3241
Direct observation of DNA threading in flap endonuclease complexes
Abstract
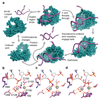
Maintenance of genome integrity requires that branched nucleic acid molecules be accurately processed to produce double-helical DNA. Flap endonucleases are essential enzymes that trim such branched molecules generated by Okazaki-fragment synthesis during replication. Here, we report crystal structures of bacteriophage T5 flap endonuclease in complexes with intact DNA substrates and products, at resolutions of 1.9-2.2 Å. They reveal single-stranded DNA threading through a hole in the enzyme, which is enclosed by an inverted V-shaped helical arch straddling the active site. Residues lining the hole induce an unusual barb-like conformation in the DNA substrate, thereby juxtaposing the scissile phosphate and essential catalytic metal ions. A series of complexes and biochemical analyses show how the substrate's single-stranded branch approaches, threads through and finally emerges on the far side of the enzyme. Our studies suggest that substrate recognition involves an unusual 'fly-casting, thread, bend and barb' mechanism.
Figures






References
-
- Zechner EL, Wu CA, Marians KJ. Coordinated leading– and lagging–strand synthesis at the Escherichia coli DNA replication fork. III. A polymerase–primase interaction governs primer size. J Biol Chem. 1992;267:4054–4063. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

