Small Traditional Human Communities Sustain Genomic Diversity over Microgeographic Scales despite Linguistic Isolation
- PMID: 27274003
- PMCID: PMC4989104
- DOI: 10.1093/molbev/msw099
Small Traditional Human Communities Sustain Genomic Diversity over Microgeographic Scales despite Linguistic Isolation
Abstract
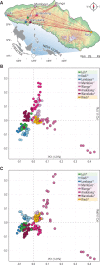
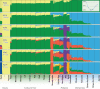
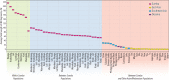
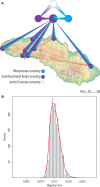
At least since the Neolithic, humans have largely lived in networks of small, traditional communities. Often socially isolated, these groups evolved distinct languages and cultures over microgeographic scales of just tens of kilometers. Population genetic theory tells us that genetic drift should act quickly in such isolated groups, thus raising the question: do networks of small human communities maintain levels of genetic diversity over microgeographic scales? This question can no longer be asked in most parts of the world, which have been heavily impacted by historical events that make traditional society structures the exception. However, such studies remain possible in parts of Island Southeast Asia and Oceania, where traditional ways of life are still practiced. We captured genome-wide genetic data, together with linguistic records, for a case-study system-eight villages distributed across Sumba, a small, remote island in eastern Indonesia. More than 4,000 years after these communities were established during the Neolithic period, most speak different languages and can be distinguished genetically. Yet their nuclear diversity is not reduced, instead being comparable to other, even much larger, regional groups. Modeling reveals a separation of time scales: while languages and culture can evolve quickly, creating social barriers, sporadic migration averaged over many generations is sufficient to keep villages linked genetically. This loosely-connected network structure, once the global norm and still extant on Sumba today, provides a living proxy to explore fine-scale genome dynamics in the sort of small traditional communities within which the most recent episodes of human evolution occurred.
Keywords: gene flow; genetic diversity; linguistic diversity; population structure..
© The Author 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
References
-
- Badan Pusat Statistik. 2010. Sensus Penduduk 2010. [cited 2016 May 20]. Available from: http://sp2010.bps.go.id
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

