Loss of function of Ywhah in mice induces deafness and cochlear outer hair cells' degeneration
- PMID: 27275396
- PMCID: PMC4893315
- DOI: 10.1038/cddiscovery.2016.17
Loss of function of Ywhah in mice induces deafness and cochlear outer hair cells' degeneration
Abstract
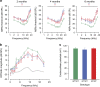
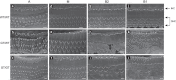
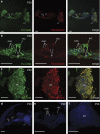
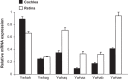
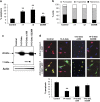
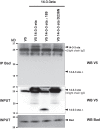
In vertebrates, 14-3-3 proteins form a family of seven highly conserved isoforms with chaperone activity, which bind phosphorylated substrates mostly involved in regulatory and checkpoint pathways. 14-3-3 proteins are the most abundant protein in the brain and are abundantly found in the cerebrospinal fluid in neurodegenerative diseases, suggesting a critical role in neuron physiology and death. Here we show that 14-3-3eta-deficient mice displayed auditory impairment accompanied by cochlear hair cells' degeneration. We show that 14-3-3eta is highly expressed in the outer and inner hair cells, spiral ganglion neurons of cochlea and retinal ganglion cells. Screening of YWHAH, the gene encoding the 14-3-3eta isoform, in non-syndromic and syndromic deafness, revealed seven non-synonymous variants never reported before. Among them, two were predicted to be damaging in families with syndromic deafness. In vitro, variants of YWHAH induce mild mitochondrial fragmentation and severe susceptibility to apoptosis, in agreement with a reduced capacity of mutated 14-3-3eta to bind the pro-apoptotic Bad protein. This study demonstrates that YWHAH variants can have a substantial effect on 14-3-3eta function and that 14-3-3eta could be a critical factor in the survival of outer hair cells.
Figures


References
-
- Ichimura T , Isobe T , Okuyama T , Yamauchi T , Fujisawa H . Brain 14-3-3 protein is an activator protein that activates tryptophan 5-monooxygenase and tyrosine 3-monooxygenase in the presence of Ca2+,calmodulin-dependent protein kinase II. FEBS Lett 1987; 219: 79–82. - PubMed
-
- Muslin AJ , Tanner JW , Allen PM , Shaw AS . Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 1996; 84: 889–897. - PubMed
-
- Yaffe MB , Rittinger K , Volinia S , Caron PR , Aitken A , Leffers H et al. The structural basis for 14-3-3: phosphopeptide binding specificity. Cell 1997; 91: 961–971. - PubMed
-
- Masters SC , Subramanian RR , Truong A , Yang H , Fujii K , Zhang H et al. Survival-promoting functions of 14-3-3 proteins. Biochem Soc Trans 2002; 30: 360–365. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

