Small Molecule Recognition and Tools to Study Modulation of r(CGG)(exp) in Fragile X-Associated Tremor Ataxia Syndrome
- PMID: 27276216
- PMCID: PMC5549791
- DOI: 10.1021/acschembio.6b00147
Small Molecule Recognition and Tools to Study Modulation of r(CGG)(exp) in Fragile X-Associated Tremor Ataxia Syndrome
Abstract
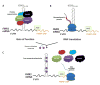
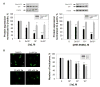
RNA transcripts containing expanded nucleotide repeats cause many incurable diseases via various mechanisms. One such disorder, fragile X-associated tremor ataxia syndrome (FXTAS), is caused by a noncoding r(CGG) repeat expansion (r(CGG)(exp)) that (i) sequesters proteins involved in RNA metabolism in nuclear foci, causing dysregulation of alternative pre-mRNA splicing, and (ii) undergoes repeat associated non-ATG translation (RANT), which produces toxic homopolymeric proteins without using a start codon. Here, we describe the design of two small molecules that inhibit both modes of toxicity and the implementation of various tools to study perturbation of these cellular events. Competitive Chemical Cross Linking and Isolation by Pull Down (C-Chem-CLIP) established that compounds bind r(CGG)(exp) and defined small molecule occupancy of r(CGG)(exp) in cells, the first approach to do so. Using an RNA GFP mimic, r(CGG)(exp)-Spinach2, we observe that our optimal designed compound binds r(CGG)(exp) and affects RNA localization by disrupting preformed RNA foci. These events correlate with an improvement of pre-mRNA splicing defects caused by RNA gain of function. In addition, the compounds reduced levels of toxic homopolymeric proteins formed via RANT. Polysome profiling studies showed that small molecules decreased loading of polysomes onto r(CGG)(exp), explaining decreased translation.
Figures





References
-
- Atkins JF, Gesteland RF, Cech TR. RNA Worlds: From Life's Orignis to Diversity in Gene Regulation. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2011.
-
- Pushechnikov A, Lee MM, Childs-Disney JL, Sobczak K, French JM, Thornton CA, Disney MD. Rational design of ligands targeting triplet repeating transcripts that cause RNA dominant disease: application to myotonic muscular dystrophy type 1 and spinocerebellar ataxia type 3. J Am Chem Soc. 2009;131:9767–9779. doi: 10.1021/ja9020149. - DOI - PMC - PubMed
-
- Coffey SM, Cook K, Tartaglia N, Tassone F, Nguyen DV, Pan R, Bronsky HE, Yuhas J, Borodyanskaya M, Grigsby J, Doerflinger M, Hagerman PJ, Hagerman RJ. Expanded clinical phenotype of women with the FMR1 premutation. Am J Med Genet Part A. 2008;146A:1009–1016. doi: 10.1002/ajmg.a.32060. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

