Genomic Classification and Prognosis in Acute Myeloid Leukemia
- PMID: 27276561
- PMCID: PMC4979995
- DOI: 10.1056/NEJMoa1516192
Genomic Classification and Prognosis in Acute Myeloid Leukemia
Abstract
Background: Recent studies have provided a detailed census of genes that are mutated in acute myeloid leukemia (AML). Our next challenge is to understand how this genetic diversity defines the pathophysiology of AML and informs clinical practice.
Methods: We enrolled a total of 1540 patients in three prospective trials of intensive therapy. Combining driver mutations in 111 cancer genes with cytogenetic and clinical data, we defined AML genomic subgroups and their relevance to clinical outcomes.
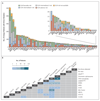
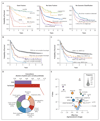
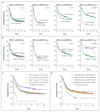
Results: We identified 5234 driver mutations across 76 genes or genomic regions, with 2 or more drivers identified in 86% of the patients. Patterns of co-mutation compartmentalized the cohort into 11 classes, each with distinct diagnostic features and clinical outcomes. In addition to currently defined AML subgroups, three heterogeneous genomic categories emerged: AML with mutations in genes encoding chromatin, RNA-splicing regulators, or both (in 18% of patients); AML with TP53 mutations, chromosomal aneuploidies, or both (in 13%); and, provisionally, AML with IDH2(R172) mutations (in 1%). Patients with chromatin-spliceosome and TP53-aneuploidy AML had poor outcomes, with the various class-defining mutations contributing independently and additively to the outcome. In addition to class-defining lesions, other co-occurring driver mutations also had a substantial effect on overall survival. The prognostic effects of individual mutations were often significantly altered by the presence or absence of other driver mutations. Such gene-gene interactions were especially pronounced for NPM1-mutated AML, in which patterns of co-mutation identified groups with a favorable or adverse prognosis. These predictions require validation in prospective clinical trials.
Conclusions: The driver landscape in AML reveals distinct molecular subgroups that reflect discrete paths in the evolution of AML, informing disease classification and prognostic stratification. (Funded by the Wellcome Trust and others; ClinicalTrials.gov number, NCT00146120.).
Figures

Comment in
-
Roads Diverge--A Robert Frost View of Leukemia Development.N Engl J Med. 2016 Jun 9;374(23):2282-4. doi: 10.1056/NEJMe1603420. N Engl J Med. 2016. PMID: 27276567 No abstract available.
-
Genomic Classification in Acute Myeloid Leukemia.N Engl J Med. 2016 Sep 1;375(9):900-1. doi: 10.1056/NEJMc1608739. N Engl J Med. 2016. PMID: 27579651 No abstract available.
-
Genomic Classification in Acute Myeloid Leukemia.N Engl J Med. 2016 Sep 1;375(9):900. doi: 10.1056/NEJMc1608739. N Engl J Med. 2016. PMID: 27579652 No abstract available.
-
Genomic classification of acute myeloid leukaemia: An incessantly evolving concept.Natl Med J India. 2016 Sep-Oct;29(5):283-285. Natl Med J India. 2016. PMID: 28098084 No abstract available.
References
-
- Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–52. - PubMed
-
- Grimwade D, Hills RK, Moorman AV, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–65. - PubMed
-
- Döhner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–74. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
