Diet may influence the oral microbiome composition in cats
- PMID: 27277498
- PMCID: PMC4899902
- DOI: 10.1186/s40168-016-0169-y
Diet may influence the oral microbiome composition in cats
Abstract
Background: Periodontal disease is highly prevalent amongst domestic cats, causing pain, gingival bleeding, reduced food intake, loss of teeth and possibly impacts on overall systemic health. Diet has been suggested to play a role in the development of periodontal disease in cats. There is a complete lack of information about how diet (composition and texture) affects the feline oral microbiome, the composition of which may influence oral health and the development of periodontal disease. We undertook a pilot study to assess if lifelong feeding of dry extruded kibble or wet (canned and/or fresh meat combinations) diets to cats (n = 10) with variable oral health affected the microbiome.
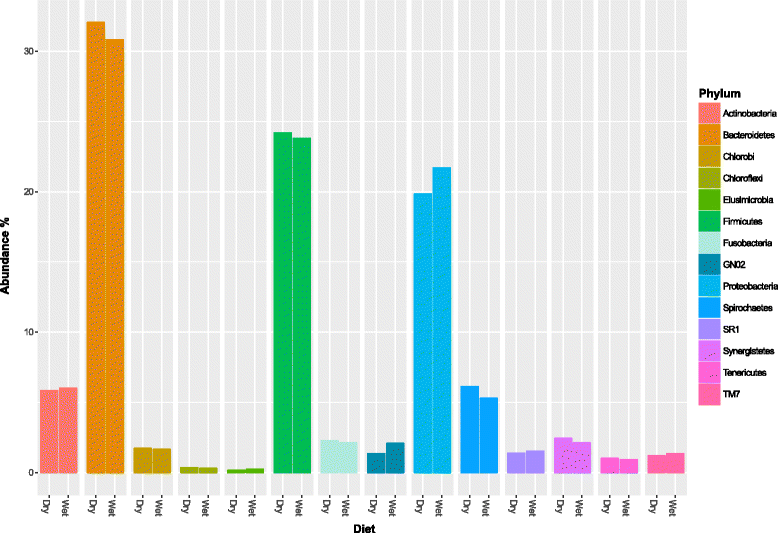
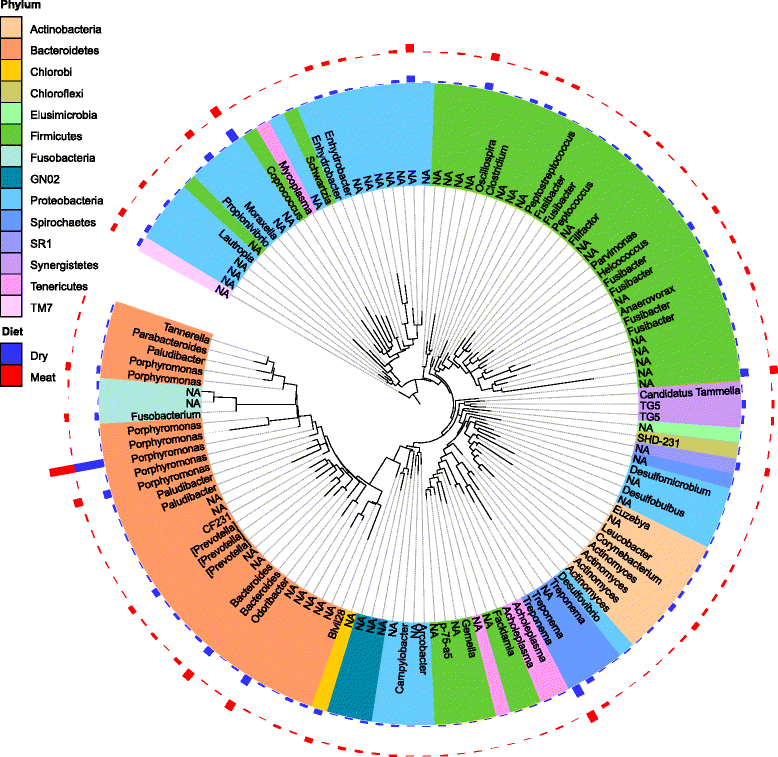
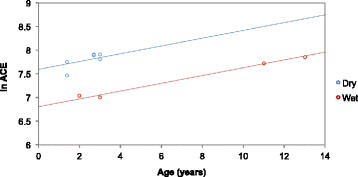
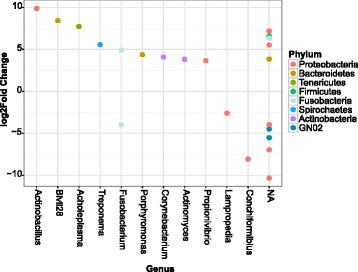
Results: Oral microbiome composition was assessed by amplifying the V1-V3 region of the 16S gene from supragingival dental plaque DNA extracts. These amplicons were sequenced using Illumina technology. This deep sequencing revealed the feline oral microbiome to be diverse, containing 411 bacterial species from 14 phyla. We found that diet had a significant influence on the overall diversity and abundance of specific bacteria in the oral environment. Cats fed a dry diet exclusively had higher bacterial diversity in their oral microbiome than wet-food diet cats (p < 0.001). Amongst this higher diversity, cats on dry-food diets had a higher abundance of Porphyromonas spp. (p < 0.01) and Treponema spp. (p < 0.01).
Conclusions: While we observed differences in the oral microbiome between cats on the two diets assessed, the relationship between these differences and gingival health was unclear. Our preliminary results indicate that further analysis of the influence of dietary constituents and texture on the feline oral microbiome is required to reveal the relationship between diet, the oral microbiome and gingival health in cats.
Keywords: 16S rRNA; Diet; Feline; Next-generation sequencing; Oral microbiome.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

