Sporozoite Route of Infection Influences In Vitro var Gene Transcription of Plasmodium falciparum Parasites From Controlled Human Infections
- PMID: 27279526
- PMCID: PMC4996147
- DOI: 10.1093/infdis/jiw225
Sporozoite Route of Infection Influences In Vitro var Gene Transcription of Plasmodium falciparum Parasites From Controlled Human Infections
Abstract
Background: Antigenic variation in Plasmodium falciparum is mediated by the multicopy var gene family. Each parasite possesses about 60 var genes, and switching between active var loci results in antigenic variation. In the current study, the effect of mosquito and host passage on in vitro var gene transcription was investigated.
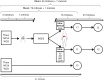
Methods: Thirty malaria-naive individuals were inoculated by intradermal or intravenous injection with cryopreserved, isogenic NF54 P. falciparum sporozoites (PfSPZ) generated from 1 premosquito culture. Microscopic parasitemia developed in 22 individuals, and 21 in vitro cultures were established. The var gene transcript levels were determined in early and late postpatient cultures and in the premosquito culture.
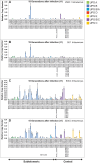
Results: At the early time point, all cultures preferentially transcribed 8 subtelomeric var genes. Intradermal infections had higher var gene transcript levels than intravenous infections and a significantly longer intrahost replication time (P = .03). At the late time point, 9 subtelomeric and 8 central var genes were transcribed at the same levels in almost all cultures. Premosquito and late postpatient cultures transcribed the same subtelomeric and central var genes, except for var2csa
Conclusions: The duration of intrahost replication influences in vitro var gene transcript patterns. Differences between premosquito and postpatient cultures decrease with prolonged in vitro growth.
Keywords: Plasmodium falciparum; controlled human malaria infections; epigenetics; malaria; transcription; var genes.
© The Author 2016. Published by Oxford University Press for the Infectious Diseases Society of America. All rights reserved. For permissions, e-mail journals.permissions@oup.com.
Figures




References
-
- World Health Organization. World malaria report 2014. Available at: http://www.who.int/malaria/publications/world_malaria_report_2014/report... Accessed 27 March 2015.
-
- Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature 2002; 415:673–9. - PubMed
-
- Su XZ, Heatwole VM, Wertheimer SP et al. . The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 1995; 82:89–100. - PubMed
-
- Baruch DI, Pasloske BL, Singh HB et al. . Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell 1995; 82:77–87. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

