Arabidopsis RNA Polymerases IV and V Are Required To Establish H3K9 Methylation, but Not Cytosine Methylation, on Geminivirus Chromatin
- PMID: 27279611
- PMCID: PMC4984644
- DOI: 10.1128/JVI.00656-16
Arabidopsis RNA Polymerases IV and V Are Required To Establish H3K9 Methylation, but Not Cytosine Methylation, on Geminivirus Chromatin
Abstract
In plants, RNA-directed DNA methylation (RdDM) employs small RNAs to target enzymes that methylate cytosine residues. Cytosine methylation and dimethylation of histone 3 lysine 9 (H3K9me2) are often linked. Together they condition an epigenetic defense that results in chromatin compaction and transcriptional silencing of transposons and viral chromatin. Canonical RdDM (Pol IV-RdDM), involving RNA polymerases IV and V (Pol IV and Pol V), was believed to be necessary to establish cytosine methylation, which in turn could recruit H3K9 methyltransferases. However, recent studies have revealed that a pathway involving Pol II and RNA-dependent RNA polymerase 6 (RDR6) (RDR6-RdDM) is likely responsible for establishing cytosine methylation at naive loci, while Pol IV-RdDM acts to reinforce and maintain it. We used the geminivirus Beet curly top virus (BCTV) as a model to examine the roles of Pol IV and Pol V in establishing repressive viral chromatin methylation. As geminivirus chromatin is formed de novo in infected cells, these viruses are unique models for processes involved in the establishment of epigenetic marks. We confirm that Pol IV and Pol V are not needed to establish viral DNA methylation but are essential for its amplification. Remarkably, however, both Pol IV and Pol V are required for deposition of H3K9me2 on viral chromatin. Our findings suggest that cytosine methylation alone is not sufficient to trigger de novo deposition of H3K9me2 and further that Pol IV-RdDM is responsible for recruiting H3K9 methyltransferases to viral chromatin.
Importance: In plants, RNA-directed DNA methylation (RdDM) uses small RNAs to target cytosine methylation, which is often linked to H3K9me2. These epigenetic marks silence transposable elements and DNA virus genomes, but how they are established is not well understood. Canonical RdDM, involving Pol IV and Pol V, was thought to establish cytosine methylation that in turn could recruit H3K9 methyltransferases, but recent studies compel a reevaluation of this view. We used BCTV to investigate the roles of Pol IV and Pol V in chromatin methylation. We found that both are needed to amplify, but not to establish, DNA methylation. However, both are required for deposition of H3K9me2. Our findings suggest that cytosine methylation is not sufficient to recruit H3K9 methyltransferases to naive viral chromatin and further that Pol IV-RdDM is responsible.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
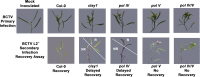
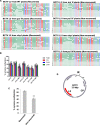
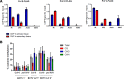

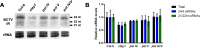
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

