Hyperactivation of ATM upon DNA-PKcs inhibition modulates p53 dynamics and cell fate in response to DNA damage
- PMID: 27280387
- PMCID: PMC4966978
- DOI: 10.1091/mbc.E16-01-0032
Hyperactivation of ATM upon DNA-PKcs inhibition modulates p53 dynamics and cell fate in response to DNA damage
Abstract
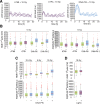
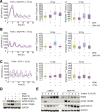
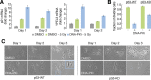
A functional DNA damage response is essential for maintaining genome integrity in the presence of DNA double-strand breaks. It is mainly coordinated by the kinases ATM, ATR, and DNA-PKcs, which control the repair of broken DNA strands and relay the damage signal to the tumor suppressor p53 to induce cell cycle arrest, apoptosis, or senescence. Although many functions of the individual kinases have been identified, it remains unclear how they act in concert to ensure faithful processing of the damage signal. Using specific inhibitors and quantitative analysis at the single-cell level, we systematically characterize the contribution of each kinase for regulating p53 activity. Our results reveal a new regulatory interplay in which loss of DNA-PKcs function leads to hyperactivation of ATM and amplification of the p53 response, sensitizing cells for damage-induced senescence. This interplay determines the outcome of treatment regimens combining irradiation with DNA-PKcs inhibitors in a p53-dependent manner.
© 2016 Finzel et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures

References
-
- Borcherds W, Theillet F-X, Katzer A, Finzel A, Mishall KM, Powell AT, Wu H, Manieri W, Dieterich C, Selenko P, et al. Disorder and residual helicity alter p53-Mdm2 binding affinity and signaling in cells. Nat Chem Biol. 2014;10:1000–1002. - PubMed
-
- Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

