Sequential Dysfunction and Progressive Depletion of Candida albicans-Specific CD4 T Cell Response in HIV-1 Infection
- PMID: 27280548
- PMCID: PMC4900544
- DOI: 10.1371/journal.ppat.1005663
Sequential Dysfunction and Progressive Depletion of Candida albicans-Specific CD4 T Cell Response in HIV-1 Infection
Abstract
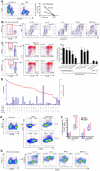
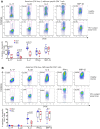
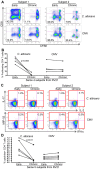
Loss of immune control over opportunistic infections can occur at different stages of HIV-1 (HIV) disease, among which mucosal candidiasis caused by the fungal pathogen Candida albicans (C. albicans) is one of the early and common manifestations in HIV-infected human subjects. The underlying immunological basis is not well defined. We have previously shown that compared to cytomegalovirus (CMV)-specific CD4 cells, C. albicans-specific CD4 T cells are highly permissive to HIV in vitro. Here, based on an antiretroviral treatment (ART) naïve HIV infection cohort (RV21), we investigated longitudinally the impact of HIV on C. albicans- and CMV-specific CD4 T-cell immunity in vivo. We found a sequential dysfunction and preferential depletion for C. albicans-specific CD4 T cell response during progressive HIV infection. Compared to Th1 (IFN-γ, MIP-1β) functional subsets, the Th17 functional subsets (IL-17, IL-22) of C. albicans-specific CD4 T cells were more permissive to HIV in vitro and impaired earlier in HIV-infected subjects. Infection history analysis showed that C. albicans-specific CD4 T cells were more susceptible to HIV in vivo, harboring modestly but significantly higher levels of HIV DNA, than CMV-specific CD4 T cells. Longitudinal analysis of HIV-infected individuals with ongoing CD4 depletion demonstrated that C. albicans-specific CD4 T-cell response was preferentially and progressively depleted. Taken together, these data suggest a potential mechanism for earlier loss of immune control over mucosal candidiasis in HIV-infected patients and provide new insights into pathogen-specific immune failure in AIDS pathogenesis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Munoz A, Schrager LK, Bacellar H, Speizer I, Vermund SH, Detels R, et al. Trends in the incidence of outcomes defining acquired immunodeficiency syndrome (AIDS) in the Multicenter AIDS Cohort Study: 1985–1991. Am J Epidemiol. 1993;137(4):423–38. . - PubMed
-
- Mocroft A, Youle M, Phillips AN, Halai R, Easterbrook P, Johnson MA, et al. The incidence of AIDS-defining illnesses in 4883 patients with human immunodeficiency virus infection. Royal Free/Chelsea and Westminster Hospitals Collaborative Group. Arch Intern Med. 1998;158(5):491–7. . - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

