IFI16 Preferentially Binds to DNA with Quadruplex Structure and Enhances DNA Quadruplex Formation
- PMID: 27280708
- PMCID: PMC4900677
- DOI: 10.1371/journal.pone.0157156
IFI16 Preferentially Binds to DNA with Quadruplex Structure and Enhances DNA Quadruplex Formation
Abstract
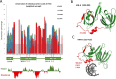
Interferon-inducible protein 16 (IFI16) is a member of the HIN-200 protein family, containing two HIN domains and one PYRIN domain. IFI16 acts as a sensor of viral and bacterial DNA and is important for innate immune responses. IFI16 binds DNA and binding has been described to be DNA length-dependent, but a preference for supercoiled DNA has also been demonstrated. Here we report a specific preference of IFI16 for binding to quadruplex DNA compared to other DNA structures. IFI16 binds to quadruplex DNA with significantly higher affinity than to the same sequence in double stranded DNA. By circular dichroism (CD) spectroscopy we also demonstrated the ability of IFI16 to stabilize quadruplex structures with quadruplex-forming oligonucleotides derived from human telomere (HTEL) sequences and the MYC promotor. A novel H/D exchange mass spectrometry approach was developed to assess protein interactions with quadruplex DNA. Quadruplex DNA changed the IFI16 deuteration profile in parts of the PYRIN domain (aa 0-80) and in structurally identical parts of both HIN domains (aa 271-302 and aa 586-617) compared to single stranded or double stranded DNAs, supporting the preferential affinity of IFI16 for structured DNA. Our results reveal the importance of quadruplex DNA structure in IFI16 binding and improve our understanding of how IFI16 senses DNA. IFI16 selectivity for quadruplex structure provides a mechanistic framework for IFI16 in immunity and cellular processes including DNA damage responses and cell proliferation.
Conflict of interest statement
Figures


Similar articles
-
New insights into the structural basis of DNA recognition by HINa and HINb domains of IFI16.J Mol Cell Biol. 2016 Feb;8(1):51-61. doi: 10.1093/jmcb/mjv053. Epub 2015 Aug 5. J Mol Cell Biol. 2016. PMID: 26246511
-
Bacterial expression of the HINab domain of IFI16: purification, characterization of DNA binding activity, and co-crystallization with viral dsDNA.Protein Expr Purif. 2014 Oct;102:13-9. doi: 10.1016/j.pep.2014.07.004. Epub 2014 Jul 19. Protein Expr Purif. 2014. PMID: 25050461
-
Charge-Mediated Pyrin Oligomerization Nucleates Antiviral IFI16 Sensing of Herpesvirus DNA.mBio. 2019 Jul 23;10(4):e01428-19. doi: 10.1128/mBio.01428-19. mBio. 2019. PMID: 31337724 Free PMC article.
-
Interferon-inducible p200-family protein IFI16, an innate immune sensor for cytosolic and nuclear double-stranded DNA: regulation of subcellular localization.Mol Immunol. 2012 Jan;49(4):567-71. doi: 10.1016/j.molimm.2011.11.004. Epub 2011 Dec 2. Mol Immunol. 2012. PMID: 22137500 Free PMC article. Review.
-
IFI16: At the interphase between innate DNA sensing and genome regulation.Cytokine Growth Factor Rev. 2014 Dec;25(6):649-55. doi: 10.1016/j.cytogfr.2014.06.004. Epub 2014 Jun 21. Cytokine Growth Factor Rev. 2014. PMID: 25027602 Review.
Cited by
-
Cellular Factors Targeting HIV-1 Transcription and Viral RNA Transcripts.Viruses. 2020 Apr 29;12(5):495. doi: 10.3390/v12050495. Viruses. 2020. PMID: 32365692 Free PMC article. Review.
-
In-Depth Bioinformatic Analyses of Nidovirales Including Human SARS-CoV-2, SARS-CoV, MERS-CoV Viruses Suggest Important Roles of Non-canonical Nucleic Acid Structures in Their Lifecycles.Front Microbiol. 2020 Jul 3;11:1583. doi: 10.3389/fmicb.2020.01583. eCollection 2020. Front Microbiol. 2020. PMID: 32719673 Free PMC article.
-
Amino Acid Composition in Various Types of Nucleic Acid-Binding Proteins.Int J Mol Sci. 2021 Jan 18;22(2):922. doi: 10.3390/ijms22020922. Int J Mol Sci. 2021. PMID: 33477647 Free PMC article. Review.
-
The dual roles of human PYHIN family proteins in cancer: mechanisms and therapeutic implications.Front Immunol. 2025 May 2;16:1576674. doi: 10.3389/fimmu.2025.1576674. eCollection 2025. Front Immunol. 2025. PMID: 40386782 Free PMC article. Review.
-
Interferon-Inducible Protein 16 (IFI16) Has a Broad-Spectrum Binding Ability Against ssDNA Targets: An Evolutionary Hypothesis for Antiretroviral Checkpoint.Front Microbiol. 2019 Jul 4;10:1426. doi: 10.3389/fmicb.2019.01426. eCollection 2019. Front Microbiol. 2019. PMID: 31333597 Free PMC article.
References
-
- Singh VV, Kerur N, Bottero V, Dutta S, Chakraborty S, Ansari MA, et al. Kaposi's sarcoma-associated herpesvirus latency in endothelial and B cells activates gamma interferon-inducible protein 16-mediated inflammasomes. J Virol. 2013;87(8):4417–31. Epub 2013/02/08. 10.1128/JVI.03282-12 - DOI - PMC - PubMed
-
- Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, et al. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9(5):363–75. Epub 2011/05/18. 10.1016/j.chom.2011.04.008 S1931-3128(11)00130-2 [pii]. - DOI - PMC - PubMed
-
- Johnstone RW, Kerry JA, Trapani JA. The human interferon-inducible protein, IFI 16, is a repressor of transcription. J Biol Chem. 1998;273(27):17172–7. Epub 1998/06/27. . - PubMed
-
- Thompson MR, Sharma S, Atianand M, Jensen SB, Carpenter S, Knipe DM, et al. Interferon gamma-inducible protein (IFI) 16 transcriptionally regulates type i interferons and other interferon-stimulated genes and controls the interferon response to both DNA and RNA viruses. J Biol Chem. 2014;289(34):23568–81. Epub 2014/07/09. 10.1074/jbc.M114.554147 M114.554147 [pii]. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

