Wnt-beta-catenin pathway signals metastasis-associated tumor cell phenotypes in triple negative breast cancers
- PMID: 27281609
- PMCID: PMC5190013
- DOI: 10.18632/oncotarget.8988
Wnt-beta-catenin pathway signals metastasis-associated tumor cell phenotypes in triple negative breast cancers
Abstract
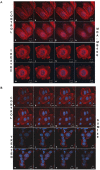
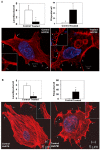
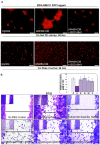
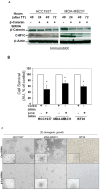
Tumor cells acquire metastasis-associated (MA) phenotypes following genetic alterations in them which cause deregulation of different signaling pathways. Earlier, we reported that an upregulation of the Wnt-beta-catenin pathway (WP) is one of the genetic salient features of triple-negative breast cancer (TNBC), and WP signaling is associated with metastasis in TNBC. Using cBioPortal, here we found that collective % of alteration(s) in WP genes, CTNNB1, APC and DVL1 among breast-invasive-carcinomas was 21% as compared to 56% in PAM50 Basal. To understand the functional relevance of WP in the biology of heterogeneous/metastasizing TNBC cells, we undertook this comprehensive study using 15 cell lines in which we examined the role of WP in the context of integrin-dependent MA-phenotypes. Directional movement of tumor cells was observed by confocal immunofluorescence microscopy and quantitative confocal-video-microscopy while matrigel-invasion was studied by MMP7-specific casein-zymography. WntC59, XAV939, sulindac sulfide and beta-catenin siRNA (1) inhibited fibronectin-directed migration, (2) decreased podia-parameters and motility-descriptors, (3) altered filamentous-actin, (4) decreased matrigel-invasion and (5) inhibited cell proliferation as well as 3D clonogenic growth. Sulindac sulfide and beta-catenin siRNA decreased beta-catenin/active-beta-catenin and MMP7. LWnt3ACM-stimulated proliferation, clonogenicity, fibronectin-directed migration and matrigel-invasion were perturbed by WP-modulators, sulindac sulfide and GDC-0941. We studied a direct involvement of WP in metastasis by stimulating brain-metastasis-specific MDA-MB231BR cells to demonstrate that LWnt3ACM-stimulated proliferation, clonogenicity and migration were blocked following sulindac sulfide, GDC-0941 and beta-catenin knockdown. We present the first evidence showing a direct functional relationship between WP activation and integrin-dependent MA-phenotypes. By proving the functional relationship between WP activation and MA-phenotypes, our data mechanistically explains (1) why different components of WP are upregulated in TNBC, (2) how WP activation is associated with metastasis and (3) how integrin-dependent MA-phenotypes can be regulated by mitigating the WP.
Keywords: LWnt3ACM stimulation; Wnt-pathway modulators; brain-metastasis specific TNBC cells; integrin-directed migration; matrigel-invasion.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures







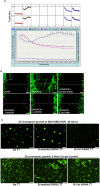
References
-
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. - PubMed
-
- Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nature reviews Clinical oncology. 2010;7:683–692. - PubMed
-
- Reis-Filho JS, Tutt AN. Triple negative tumours: a critical review. Histopathology. 2008;52:108–118. - PubMed
-
- Dent R, Hanna WM, Trudeau M, Rawlinson E, Sun P, Narod SA. Pattern of metastatic spread in triple-negative breast cancer. Breast cancer research and treatment. 2009;115:423–428. - PubMed
-
- Carey LA. Directed therapy of subtypes of triple-negative breast cancer. The oncologist. 2011;16(Suppl 1):71–78. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

