Initial Segment Differentiation Begins During a Critical Window and Is Dependent upon Lumicrine Factors and SRC Proto-Oncogene (SRC) in the Mouse
- PMID: 27281706
- PMCID: PMC5029432
- DOI: 10.1095/biolreprod.116.138388
Initial Segment Differentiation Begins During a Critical Window and Is Dependent upon Lumicrine Factors and SRC Proto-Oncogene (SRC) in the Mouse
Abstract
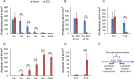
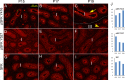
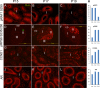
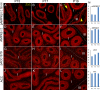
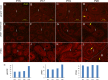
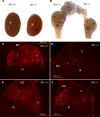
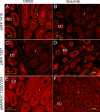
Without a fully developed and functioning initial segment, the most proximal region of the epididymis, male infertility results. Therefore, it is important to understand the development of the initial segment. During postnatal development of the epididymis, many cellular processes of the initial segment are regulated by lumicrine factors, which are produced by the testis and enter the epididymis with testicular luminal fluid. In this report, we showed that prior to Postnatal Day 15 (P15), the initial segment was lumicrine factor independent in the mouse. However, from P19 onward, lumicrine factors were essential for the proliferation and survival of initial segment epithelial cells. Therefore, P15 to P19 was a critical window that established the dependency of lumicrine factors in the initial segment epithelium. The initial segment-specific kinase activity profile, a marker of initial segment differentiation, was also established during this window. The SFK (SRC proto-oncogene family kinases), ERK pathway (known as the RAF/MEK/ERK pathway) components, and AMPK (AMP-activated protein kinases) pathway components had increased activities from P15 to P19, suggesting that lumicrine factors regulated SFK/ERK/AMPK signaling to initiate differentiation of the initial segment from P15 to P19. Compared with litter mate controls, juvenile Src null mice displayed lower levels of MAPK3/1 (mitogen-activated protein kinase 3/1) activity and a reduced level of differentiation in the initial segment epithelium, a similar phenotype resulting from inhibition of SRC activity within the window of P15 to P19. Therefore, lumicrine factor-dependent SRC activity signaling through MAPK3/1 is important for the initiation of initial segment differentiation during a critical window of development.
Keywords: ERK pathway; SRC; differentiation; epididymis; lumicrine regulation.
© 2016 by the Society for the Study of Reproduction, Inc.
Figures
References
-
- Hinton BT, Galdamez MM, Sutherland A, Bomgardner D, Xu B, Abdel-Fattah R, Yang L. How do you get six meters of epididymis inside a human scrotum? J Androl. 2011;32:558–564. - PubMed
-
- Robaire B, Hinton BT. The epididymis. In: Plant T, Zeleznik AJ, editors. Knobil and Neill's Physiology of Reproduction, 4th ed. New York: Elsevier; 2014. pp. 691–772. In. (eds.)
-
- Jelinsky SA, Turner TT, Bang HJ, Finger JN, Solarz MK, Wilson E, Brown EL, Kopf GS, Johnston DS. The rat epididymal transcriptome: comparison of segmental gene expression in the rat and mouse epididymides. Biol Reprod. 2007;76:561–570. - PubMed
-
- Johnston DS, Turner TT, Finger JN, Owtscharuk TL, Kopf GS, Jelinsky SA. Identification of epididymis-specific transcripts in the mouse and rat by transcriptional profiling. Asian J Androl. 2007;9:522–527. - PubMed
-
- Sonnenberg-Riethmacher E, Walter B, Riethmacher D, Godecke S, Birchmeier C. The c-ros tyrosine kinase receptor controls regionalization and differentiation of epithelial cells in the epididymis. Genes Dev. 1996;10:1184–1193. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

