Genome-wide detection of CNVs in Chinese indigenous sheep with different types of tails using ovine high-density 600K SNP arrays
- PMID: 27282145
- PMCID: PMC4901276
- DOI: 10.1038/srep27822
Genome-wide detection of CNVs in Chinese indigenous sheep with different types of tails using ovine high-density 600K SNP arrays
Abstract
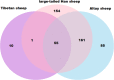
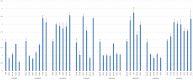
Chinese indigenous sheep can be classified into three types based on tail morphology: fat-tailed, fat-rumped, and thin-tailed sheep, of which the typical breeds are large-tailed Han sheep, Altay sheep, and Tibetan sheep, respectively. To unravel the genetic mechanisms underlying the phenotypic differences among Chinese indigenous sheep with tails of three different types, we used ovine high-density 600K SNP arrays to detect genome-wide copy number variation (CNV). In large-tailed Han sheep, Altay sheep, and Tibetan sheep, 371, 301, and 66 CNV regions (CNVRs) with lengths of 71.35 Mb, 51.65 Mb, and 10.56 Mb, respectively, were identified on autosomal chromosomes. Ten CNVRs were randomly chosen for confirmation, of which eight were successfully validated. The detected CNVRs harboured 3130 genes, including genes associated with fat deposition, such as PPARA, RXRA, KLF11, ADD1, FASN, PPP1CA, PDGFA, and PEX6. Moreover, multilevel bioinformatics analyses of the detected candidate genes were significantly enriched for involvement in fat deposition, GTPase regulator, and peptide receptor activities. This is the first high-resolution sheep CNV map for Chinese indigenous sheep breeds with three types of tails. Our results provide valuable information that will support investigations of genomic structural variation underlying traits of interest in sheep.
Figures
References
-
- Safdarian M., Zamiri M. J., Hashemi M. & Noorolahi H. Relationships of fat-tail dimensions with fat-tail weight and carcass characteristics at different slaughter weights of Torki-Ghashghaii sheep. Meat Sci. 80, 686–689 (2008). - PubMed
-
- Ermias E., Yami A. & Rege J. Fat deposition in tropical sheep as adaptive attribute to periodic feed fluctuation. J. Anim. Breed. Genet. 119, 235–246 (2002).
-
- Kashan N. E. J., Azar G. H. M., Afzalzadeh A. & Salehi A. Growth performance and carcass quality of fattening lambs from fat-tailed and tailed sheep breeds. Small Ruminant Res. 60, 267–271 (2005).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

