Architecture and RNA binding of the human negative elongation factor
- PMID: 27282391
- PMCID: PMC4940160
- DOI: 10.7554/eLife.14981
Architecture and RNA binding of the human negative elongation factor
Abstract
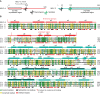
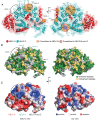
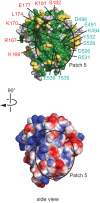
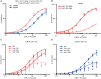
Transcription regulation in metazoans often involves promoter-proximal pausing of RNA polymerase (Pol) II, which requires the 4-subunit negative elongation factor (NELF). Here we discern the functional architecture of human NELF through X-ray crystallography, protein crosslinking, biochemical assays, and RNA crosslinking in cells. We identify a NELF core subcomplex formed by conserved regions in subunits NELF-A and NELF-C, and resolve its crystal structure. The NELF-AC subcomplex binds single-stranded nucleic acids in vitro, and NELF-C associates with RNA in vivo. A positively charged face of NELF-AC is involved in RNA binding, whereas the opposite face of the NELF-AC subcomplex binds NELF-B. NELF-B is predicted to form a HEAT repeat fold, also binds RNA in vivo, and anchors the subunit NELF-E, which is confirmed to bind RNA in vivo. These results reveal the three-dimensional architecture and three RNA-binding faces of NELF.
Keywords: RNA polymerase II; biochemistry; biophysics; gene regulation; gene transcription; human; promoter-proximal pausing; structural biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures









Similar articles
-
Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex.Proc Natl Acad Sci U S A. 2010 Jun 22;107(25):11301-6. doi: 10.1073/pnas.1000681107. Epub 2010 Jun 4. Proc Natl Acad Sci U S A. 2010. PMID: 20534440 Free PMC article.
-
Structural studies on the RNA-recognition motif of NELF E, a cellular negative transcription elongation factor involved in the regulation of HIV transcription.Biochem J. 2006 Dec 15;400(3):449-56. doi: 10.1042/BJ20060421. Biochem J. 2006. PMID: 16898873 Free PMC article.
-
Structure of paused transcription complex Pol II-DSIF-NELF.Nature. 2018 Aug;560(7720):601-606. doi: 10.1038/s41586-018-0442-2. Epub 2018 Aug 22. Nature. 2018. PMID: 30135580 Free PMC article.
-
Regulation of Promoter Proximal Pausing of RNA Polymerase II in Metazoans.J Mol Biol. 2021 Jul 9;433(14):166897. doi: 10.1016/j.jmb.2021.166897. Epub 2021 Feb 25. J Mol Biol. 2021. PMID: 33640324 Free PMC article. Review.
-
Transcription elongation factors DSIF and NELF: promoter-proximal pausing and beyond.Biochim Biophys Acta. 2013 Jan;1829(1):98-104. doi: 10.1016/j.bbagrm.2012.11.007. Epub 2012 Nov 29. Biochim Biophys Acta. 2013. PMID: 23202475 Review.
Cited by
-
RNA Polymerase II "Pause" Prepares Promoters for Upcoming Transcription during Drosophila Development.Int J Mol Sci. 2022 Sep 13;23(18):10662. doi: 10.3390/ijms231810662. Int J Mol Sci. 2022. PMID: 36142573 Free PMC article.
-
Constrained G4 structures unveil topology specificity of known and new G4 binding proteins.Sci Rep. 2021 Jun 29;11(1):13469. doi: 10.1038/s41598-021-92806-8. Sci Rep. 2021. PMID: 34188089 Free PMC article.
-
Structural basis for mutually exclusive co-transcriptional nuclear cap-binding complexes with either NELF-E or ARS2.Nat Commun. 2017 Nov 3;8(1):1302. doi: 10.1038/s41467-017-01402-w. Nat Commun. 2017. PMID: 29101316 Free PMC article.
-
Structural insights into promoter-proximal pausing of RNA polymerase II at +1 nucleosome.Sci Adv. 2025 Mar 7;11(10):eadu0577. doi: 10.1126/sciadv.adu0577. Epub 2025 Mar 5. Sci Adv. 2025. PMID: 40043114 Free PMC article.
-
NELFE promoted pancreatic cancer metastasis and the epithelial‑to‑mesenchymal transition by decreasing the stabilization of NDRG2 mRNA.Int J Oncol. 2019 Dec;55(6):1313-1323. doi: 10.3892/ijo.2019.4890. Epub 2019 Oct 2. Int J Oncol. 2019. PMID: 31638184 Free PMC article.
References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica. Section D, Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
-
- Aiyar SE, Sun JL, Blair AL, Moskaluk CA, Lu YZ, Ye QN, Yamaguchi Y, Mukherjee A, Ren DM, Handa H, Li R, Handa H, Li H. Attenuation of estrogen receptor alpha-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes & Development. 2004;18:2134–2146. doi: 10.1101/gad.1214104. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

