The complete genome of a baculovirus isolated from an insect of medical interest: Lonomia obliqua (Lepidoptera: Saturniidae)
- PMID: 27282807
- PMCID: PMC4901303
- DOI: 10.1038/srep23127
The complete genome of a baculovirus isolated from an insect of medical interest: Lonomia obliqua (Lepidoptera: Saturniidae)
Abstract
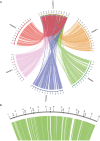
Lonomia obliqua (Lepidoptera: Saturniidae) is a species of medical importance due to the severity of reactions caused by accidental contact with the caterpillar bristles. Several natural pathogens have been identified in L. obliqua, and among them the baculovirus Lonomia obliqua multiple nucleopolyhedrovirus (LoobMNPV). The complete genome of LoobMNPV was sequenced and shown to have 120,022 bp long with 134 putative open reading frames (ORFs). Phylogenetic analysis of the LoobMNPV genome showed that it belongs to Alphabaculovirus group I (lepidopteran-infective NPV). A total of 12 unique ORFs were identified with no homologs in other sequenced baculovirus genomes. One of these, the predicted protein encoded by loob035, showed significant identity to an eukaryotic transcription terminator factor (TTF2) from the Lepidoptera Danaus plexippus, suggesting an independent acquisition through horizontal gene transfer. Homologs of cathepsin and chitinase genes, which are involved in host integument liquefaction and viral spread, were not found in this genome. As L. obliqua presents a gregarious behavior during the larvae stage the impact of this deletion might be neglectable.
Figures





References
-
- Diaz J. H. The evolving global epidemiology, syndromic classification, management, and prevention of caterpillar envenoming. Am. J. Trop. Med. Hyg. 72, 347–357 (2005). - PubMed
-
- Carrijo-Carvalho L. C. & Chudzinski-Tavassi A. M. The venom of the Lonomia caterpillar: an overview. Toxicon 49, 741–757 (2007). - PubMed
-
- Gamborgi G. P., Metcalf E. B. & Barros E. J. G. Acute renal failure provoked by toxin from caterpillars of the species Lonomia obliqua. Toxicon 47, 68–74 (2006). - PubMed
-
- Abella H. B. et al. Manual de diagnóstico e tratamento de acidentes porLonomia.20 (Centro de Informação Toxicológica-SS/RS-FEPPS, 1999).
-
- Moraes R. H. P. Identificação dos Inimigos Naturais de Lonomia obliqua Walker, 1855 (Lepidóptera: Saturniidae) e possíveis fatos determinantes do aumento de sua população. Masters Thesis, Escola Superior de Agricultura Luiz de Queiroz (2002).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

