Histidine switch controlling pH-dependent protein folding and DNA binding in a transcription factor at the core of synthetic network devices
- PMID: 27282811
- PMCID: PMC4955742
- DOI: 10.1039/c6mb00304d
Histidine switch controlling pH-dependent protein folding and DNA binding in a transcription factor at the core of synthetic network devices
Abstract
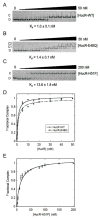
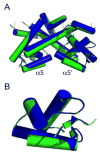
Therapeutic strategies have been reported that depend on synthetic network devices in which a urate-sensing transcriptional regulator detects pathological levels of urate and triggers production or release of urate oxidase. The transcription factor involved, HucR, is a member of the multiple antibiotic resistance (MarR) protein family. We show that protonation of stacked histidine residues at the pivot point of long helices that form the scaffold of the dimer interface leads to reversible formation of a molten globule state and significantly attenuated DNA binding at physiological temperatures. We also show that binding of urate to symmetrical sites in each protein lobe is communicated via the dimer interface. This is the first demonstration of regulation of a MarR family transcription factor by pH-dependent interconversion between a molten globule and a compact folded state. Our data further suggest that HucR may be utilized in synthetic devices that depend on detection of pH changes.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

