MicroRNAs in glioblastoma multiforme pathogenesis and therapeutics
- PMID: 27282910
- PMCID: PMC4971921
- DOI: 10.1002/cam4.775
MicroRNAs in glioblastoma multiforme pathogenesis and therapeutics
Abstract
Glioblastoma multiforme (GBM) is the most common and lethal cancer of the adult brain, remaining incurable with a median survival time of only 15 months. In an effort to identify new targets for GBM diagnostics and therapeutics, recent studies have focused on molecular phenotyping of GBM subtypes. This has resulted in mounting interest in microRNAs (miRNAs) due to their regulatory capacities in both normal development and in pathological conditions such as cancer. miRNAs have a wide range of targets, allowing them to modulate many pathways critical to cancer progression, including proliferation, cell death, metastasis, angiogenesis, and drug resistance. This review explores our current understanding of miRNAs that are differentially modulated and pathologically involved in GBM as well as the current state of miRNA-based therapeutics. As the role of miRNAs in GBM becomes more well understood and novel delivery methods are developed and optimized, miRNA-based therapies could provide a critical step forward in cancer treatment.
Keywords: cancer therapy; glioblastoma multiforme; microRNA.
© 2016 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Figures
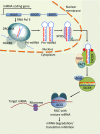
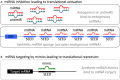
Similar articles
-
Targeting strategies on miRNA-21 and PDCD4 for glioblastoma.Arch Biochem Biophys. 2015 Aug 15;580:64-74. doi: 10.1016/j.abb.2015.07.001. Epub 2015 Jul 2. Arch Biochem Biophys. 2015. PMID: 26142886 Review.
-
Differential expression of MicroRNAs in patients with glioblastoma after concomitant chemoradiotherapy.OMICS. 2013 May;17(5):259-68. doi: 10.1089/omi.2012.0065. Epub 2013 Apr 15. OMICS. 2013. PMID: 23586679
-
The Clinical Role of miRNAs in the Development and Treatment of Glioblastoma.Int J Mol Sci. 2025 Mar 18;26(6):2723. doi: 10.3390/ijms26062723. Int J Mol Sci. 2025. PMID: 40141375 Free PMC article. Review.
-
MiRNA-21 silencing mediated by tumor-targeted nanoparticles combined with sunitinib: A new multimodal gene therapy approach for glioblastoma.J Control Release. 2015 Jun 10;207:31-9. doi: 10.1016/j.jconrel.2015.04.002. Epub 2015 Apr 7. J Control Release. 2015. PMID: 25861727
-
MicroRNAs in glioblastoma pathogenesis and therapy: A comprehensive review.Crit Rev Oncol Hematol. 2017 Dec;120:22-33. doi: 10.1016/j.critrevonc.2017.10.003. Epub 2017 Oct 7. Crit Rev Oncol Hematol. 2017. PMID: 29198335 Review.
Cited by
-
miR-205: A Potential Biomedicine for Cancer Therapy.Cells. 2020 Aug 25;9(9):1957. doi: 10.3390/cells9091957. Cells. 2020. PMID: 32854238 Free PMC article. Review.
-
MiR-148a inhibits the proliferation and migration of glioblastoma by targeting ITGA9.Hum Cell. 2019 Oct;32(4):548-556. doi: 10.1007/s13577-019-00279-9. Epub 2019 Sep 5. Hum Cell. 2019. PMID: 31489579
-
Deciphering specific miRNAs in brain tumors: a 5-miRNA signature in glioblastoma.Mol Genet Genomics. 2022 Mar;297(2):507-521. doi: 10.1007/s00438-022-01866-6. Epub 2022 Feb 17. Mol Genet Genomics. 2022. PMID: 35175428
-
Microrna-1224-5p Is a Potential Prognostic and Therapeutic Biomarker in Glioblastoma: Integrating Bioinformatics and Clinical Analyses.Curr Med Sci. 2022 Jun;42(3):584-596. doi: 10.1007/s11596-022-2593-5. Epub 2022 Jun 8. Curr Med Sci. 2022. PMID: 35678909
-
The Evaluation of Glioblastoma Cell Dissociation and Its Influence on Its Behavior.Int J Mol Sci. 2019 Sep 18;20(18):4630. doi: 10.3390/ijms20184630. Int J Mol Sci. 2019. PMID: 31540507 Free PMC article.
References
-
- Stupp, R. , Hegi M. E., Mason W. P., van den Bent M. J., Taphoorn M. J., Janzer R. C., et al. 2009. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5‐year analysis of the EORTC‐NCIC trial. Lancet Oncol. 10:459–466. - PubMed
-
- Winter, J. , Jung S., Keller S., Gregory R. I., and Diederichs S.. 2009. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 11:228–234. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

