Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut
- PMID: 27284201
- PMCID: PMC5144996
- DOI: 10.1126/science.aad9926
Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut
Abstract
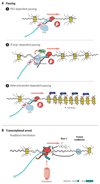
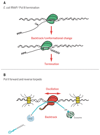
Terminating transcription is a highly intricate process for mammalian protein-coding genes. First, the chromatin template slows down transcription at the gene end. Then, the transcript is cleaved at the poly(A) signal to release the messenger RNA. The remaining transcript is selectively unraveled and degraded. This induces critical conformational changes in the heart of the enzyme that trigger termination. Termination can also occur at variable positions along the gene and so prevent aberrant transcript formation or intentionally make different transcripts. These may form multiple messenger RNAs with altered regulatory properties or encode different proteins. Finally, termination can be perturbed to achieve particular cellular needs or blocked in cancer or virally infected cells. In such cases, failure to terminate transcription can spell disaster for the cell.
Copyright © 2016, American Association for the Advancement of Science.
Figures
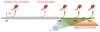

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

