Defining the Active Fraction of Daptomycin against Methicillin-Resistant Staphylococcus aureus (MRSA) Using a Pharmacokinetic and Pharmacodynamic Approach
- PMID: 27284923
- PMCID: PMC4902307
- DOI: 10.1371/journal.pone.0156131
Defining the Active Fraction of Daptomycin against Methicillin-Resistant Staphylococcus aureus (MRSA) Using a Pharmacokinetic and Pharmacodynamic Approach
Abstract
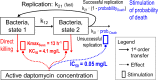
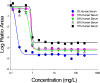
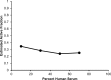
Our objective was to study the pharmacodynamics of daptomycin in the presence of varying concentrations of human serum (HS) in vitro to quantify the fraction of daptomycin that is 'active'. Time kill experiments were performed with daptomycin (0 to 256 mg/L) against two MRSA strains at log-phase growth, in the presence of HS (0%, 10%, 30%, 50%, 70%) combined with Mueller-Hinton broth. Daptomycin ≥ 2 mg/L achieved 99.9% kill within 8 h at all HS concentrations; early killing activity was slightly attenuated at higher HS concentrations. After 1 h, bacterial reduction of USA300 upon exposure to daptomycin 4 mg/L ranged from -3.1 to -0.5 log10CFU/mL in the presence of 0% to 70% HS, respectively. Bactericidal activity was achieved against both strains at daptomycin ≥ 4 mg/L for all fractions of HS exposure. A mechanism-based mathematical model (MBM) was developed to estimate the active daptomycin fraction at each %HS, comprising 3 bacterial subpopulations differing in daptomycin susceptibility. Time-kill data were fit with this MBM with excellent precision (r2 >0.95). The active fraction of daptomycin was estimated to range from 34.6% to 25.2% at HS fractions of 10% to 70%, respectively. Despite the reported low unbound fraction of daptomycin, the impact of protein binding on the activity of daptomycin was modest. The active fraction approach can be utilized to design in vitro experiments and to optimize therapeutic regimens of daptomycin in humans.
Conflict of interest statement
Figures


Similar articles
-
Evaluation of vancomycin and daptomycin against methicillin-resistant Staphylococcus aureus and heterogeneously vancomycin-intermediate S. aureus in an in vitro pharmacokinetic/pharmacodynamic model with simulated endocardial vegetations.J Antimicrob Chemother. 2009 Jan;63(1):155-60. doi: 10.1093/jac/dkn439. Epub 2008 Nov 4. J Antimicrob Chemother. 2009. PMID: 18984644
-
Influence of protein binding under controlled conditions on the bactericidal activity of daptomycin in an in vitro pharmacodynamic model.J Antimicrob Chemother. 2004 Jul;54(1):259-62. doi: 10.1093/jac/dkh259. Epub 2004 May 18. J Antimicrob Chemother. 2004. PMID: 15150172
-
Bactericidal activity of daptomycin versus vancomycin in the presence of human albumin against vancomycin-susceptible but tolerant methicillin-resistant Staphylococcus aureus (MRSA) with daptomycin minimum inhibitory concentrations of 1-2microg/mL.Int J Antimicrob Agents. 2010 Feb;35(2):131-7. doi: 10.1016/j.ijantimicag.2009.09.021. Epub 2009 Dec 16. Int J Antimicrob Agents. 2010. PMID: 20006469
-
In vitro effect of the presence of human albumin or human serum on the bactericidal activity of daptomycin against strains with the main resistance phenotypes in Gram-positives.J Antimicrob Chemother. 2007 Jun;59(6):1185-9. doi: 10.1093/jac/dkm078. Epub 2007 Apr 5. J Antimicrob Chemother. 2007. PMID: 17412725
-
Activity of simulated serum concentrations of daptomycin versus vancomycin during the first 24h of treatment in the presence of physiological albumin concentrations against vancomycin-susceptible, -tolerant or -intermediate-resistant Staphylococcus aureus.Int J Antimicrob Agents. 2011 Apr;37(4):332-8. doi: 10.1016/j.ijantimicag.2010.12.007. Int J Antimicrob Agents. 2011. PMID: 21388792
Cited by
-
The Antistaphylococcal Lysin, CF-301, Activates Key Host Factors in Human Blood To Potentiate Methicillin-Resistant Staphylococcus aureus Bacteriolysis.Antimicrob Agents Chemother. 2019 Mar 27;63(4):e02291-18. doi: 10.1128/AAC.02291-18. Print 2019 Apr. Antimicrob Agents Chemother. 2019. PMID: 30670427 Free PMC article.
-
Antimicrobial, Anti-inflammatory, and Wound Healing Properties of Myrtus communis Leaf Methanolic Extract Ointment on Burn Wound Infection Induced by Methicillin-Resistant Staphylococcus aureus in Rats.Biomed Res Int. 2024 Jun 11;2024:6758817. doi: 10.1155/2024/6758817. eCollection 2024. Biomed Res Int. 2024. PMID: 38899039 Free PMC article.
-
Clinical Pharmacokinetics of Daptomycin.Clin Pharmacokinet. 2021 Mar;60(3):271-281. doi: 10.1007/s40262-020-00968-x. Epub 2020 Dec 14. Clin Pharmacokinet. 2021. PMID: 33313994 Review.
References
-
- Wise R. The clinical relevance of protein binding and tissue concentrations in antimicrobial therapy. Clinical pharmacokinetics. 1986;11(6):470–82. Epub 1986/11/01. . - PubMed
-
- Craig WA, Welling PG. Protein binding of antimicrobials: clinical pharmacokinetic and therapeutic implications. Clinical pharmacokinetics. 1977;2(4):252–68. Epub 1977/07/01. . - PubMed
-
- Gould IM, David MZ, Esposito S, Garau J, Lina G, Mazzei T, et al. New insights into meticillin-resistant Staphylococcus aureus (MRSA) pathogenesis, treatment and resistance. International journal of antimicrobial agents. 2012;39(2):96–104. Epub 2011/12/27. 10.1016/j.ijantimicag.2011.09.028 . - DOI - PubMed
-
- Tsuji BT, von Eiff C, Kelchlin PA, Forrest A, Smith PF. Attenuated vancomycin bactericidal activity against Staphylococcus aureus hemB mutants expressing the small-colony-variant phenotype. Antimicrobial agents and chemotherapy. 2008;52(4):1533–7. Epub 2008/02/21. 10.1128/aac.01254-07 ; PubMed Central PMCID: PMCPmc2292514. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

