Brain-derived neurotrophic factor prevents dendritic retraction of adult mouse retinal ganglion cells
- PMID: 27285957
- PMCID: PMC4988502
- DOI: 10.1111/ejn.13295
Brain-derived neurotrophic factor prevents dendritic retraction of adult mouse retinal ganglion cells
Abstract
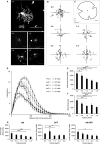
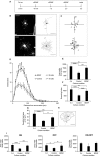
We used cultured adult mouse retinae as a model system to follow and quantify the retraction of dendrites using diolistic labelling of retinal ganglion cells (RGCs) following explantation. Cell death was monitored in parallel by nuclear staining as 'labelling' with RGC and apoptotic markers was inconsistent and exceedingly difficult to quantify reliably. Nuclear staining allowed us to delineate a lengthy time window during which dendrite retraction can be monitored in the absence of RGC death. The addition of brain-derived neurotrophic factor (BDNF) produced a marked reduction in dendritic degeneration, even when application was delayed for 3 days after retinal explantation. These results suggest that the delayed addition of trophic factors may be functionally beneficial before the loss of cell bodies in the course of conditions such as glaucoma.
Keywords: Sholl analysis; neurodegeneration; neuron labelling; neuroprotection.
© 2016 The Authors. European Journal of Neuroscience published by Federation of European Neuroscience Societies and John Wiley & Sons Ltd.
Figures





Similar articles
-
Brain-derived neurotrophic factor differentially regulates retinal ganglion cell dendritic and axonal arborization in vivo.J Neurosci. 1999 Nov 15;19(22):9928-38. doi: 10.1523/JNEUROSCI.19-22-09928.1999. J Neurosci. 1999. PMID: 10559401 Free PMC article.
-
Combined effect of brain-derived neurotrophic factor and LINGO-1 fusion protein on long-term survival of retinal ganglion cells in chronic glaucoma.Neuroscience. 2009 Aug 18;162(2):375-82. doi: 10.1016/j.neuroscience.2009.04.075. Epub 2009 May 5. Neuroscience. 2009. PMID: 19422885
-
Local and target-derived brain-derived neurotrophic factor exert opposing effects on the dendritic arborization of retinal ganglion cells in vivo.J Neurosci. 2002 Sep 1;22(17):7639-49. doi: 10.1523/JNEUROSCI.22-17-07639.2002. J Neurosci. 2002. PMID: 12196587 Free PMC article.
-
Neurotrophic factors for retinal ganglion cell neuropathy - with a special reference to diabetic neuropathy in the retina.Curr Diabetes Rev. 2014 May;10(3):166-76. doi: 10.2174/1573399810666140508121927. Curr Diabetes Rev. 2014. PMID: 24809393 Review.
-
[Advances in researches for protection and induction of cytokines to retinal neurons].Zhonghua Yan Ke Za Zhi. 2005 Sep;41(9):861-4. Zhonghua Yan Ke Za Zhi. 2005. PMID: 16191359 Review. Chinese.
Cited by
-
Gene Gun DiOlistic Labelling of Retinal Ganglion Cells.Methods Mol Biol. 2023;2708:33-40. doi: 10.1007/978-1-0716-3409-7_4. Methods Mol Biol. 2023. PMID: 37558957
-
Valproic Acid Reduces Neuroinflammation to Provide Retinal Ganglion Cell Neuroprotection in the Retina Axotomy Model.Front Cell Dev Biol. 2022 May 12;10:903436. doi: 10.3389/fcell.2022.903436. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35646919 Free PMC article.
-
Caucasian Families Exhibit Significant Linkage of Myopia to Chromosome 11p.Invest Ophthalmol Vis Sci. 2017 Jul 1;58(9):3547-3554. doi: 10.1167/iovs.16-21271. Invest Ophthalmol Vis Sci. 2017. PMID: 28715588 Free PMC article.
-
Review: Neuroprotective Nanocarriers in Glaucoma.Pharmaceuticals (Basel). 2024 Sep 10;17(9):1190. doi: 10.3390/ph17091190. Pharmaceuticals (Basel). 2024. PMID: 39338350 Free PMC article. Review.
-
Potential Therapeutic Benefit of NAD+ Supplementation for Glaucoma and Age-Related Macular Degeneration.Nutrients. 2020 Sep 19;12(9):2871. doi: 10.3390/nu12092871. Nutrients. 2020. PMID: 32961812 Free PMC article. Review.
References
-
- Bosco, A. & Linden, R. (1999) BDNF and NT‐4 differentially modulate neurite outgrowth in developing retinal ganglion cells. J. Neurosci. Res., 57, 759–769. - PubMed
-
- Caserta, T.M. , Smith, A.N. , Gultice, A.D. , Reedy, M.A. & Brown, T.L. (2003) Q‐VD‐OPh, a broad spectrum caspase inhibitor with potent antiapoptotic properties. Apoptosis, 8, 345–352. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

