Mob/oriT, a mobilizable site-specific recombination system for unmarked genetic manipulation in Bacillus thuringiensis and Bacillus cereus
- PMID: 27286821
- PMCID: PMC4902927
- DOI: 10.1186/s12934-016-0492-9
Mob/oriT, a mobilizable site-specific recombination system for unmarked genetic manipulation in Bacillus thuringiensis and Bacillus cereus
Abstract
Background: Bacillus thuringiensis and Bacillus cereus are two important species in B. cereus group. The intensive study of these strains at the molecular level and construction of genetically modified bacteria requires the development of efficient genetic tools. To insert genes into or delete genes from bacterial chromosomes, marker-less manipulation methods were employed.
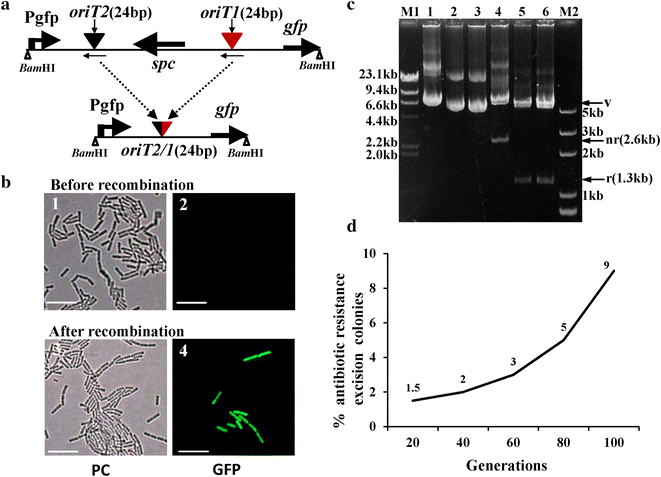
Results: We present a novel genetic manipulation method for B. thuringiensis and B. cereus strains that does not leave selection markers. Our approach takes advantage of the relaxase Mob02281 encoded by plasmid pBMB0228 from Bacillus thuringiensis. In addition to its mobilization function, this Mob protein can mediate recombination between oriT sites. The Mob02281 mobilization module was associated with a spectinomycin-resistance gene to form a Mob-Spc cassette, which was flanked by the core 24-bp oriT sequences from pBMB0228. A strain in which the wild-type chromosome was replaced with the modified copy containing the Mob-Spc cassette at the target locus was obtained via homologous recombination. Thus, the spectinomycin-resistance gene can be used to screen for Mob-Spc cassette integration mutants. Recombination between the two oriT sequences mediated by Mob02281, encoded by the Mob-Spc cassette, resulted in the excision of the Mob-Spc cassette, producing the desired chromosomal alteration without introducing unwanted selection markers. We used this system to generate an in-frame deletion of a target gene in B. thuringiensis as well as a gene located in an operon of B. cereus. Moreover, we demonstrated that this system can be used to introduce a single gene or an expression cassette of interest in B. thuringiensis.
Conclusion: The Mob/oriT recombination system provides an efficient method for unmarked genetic manipulation and for constructing genetically modified bacteria of B. thuringiensis and B. cereus. Our method extends the available genetic tools for B. thuringiensis and B. cereus strains.
Keywords: Bacillus cereus; Bacillus thuringiensis; Conjugation; Mob protein; Relaxase; Site-specific recombination.
Figures



Similar articles
-
The resolution and regeneration of a cointegrate plasmid reveals a model for plasmid evolution mediated by conjugation and oriT site-specific recombination.Environ Microbiol. 2013 Dec;15(12):3305-18. doi: 10.1111/1462-2920.12177. Epub 2013 Jul 4. Environ Microbiol. 2013. PMID: 23826996
-
Modular genetic architecture of the toxigenic plasmid pIS56-63 harboring cry1Ab21 in Bacillus thuringiensis subsp. thuringiensis strain IS5056.Pol J Microbiol. 2014;63(2):147-56. Pol J Microbiol. 2014. PMID: 25115108
-
Hemolysin II is more characteristic of Bacillus thuringiensis than Bacillus cereus.Arch Microbiol. 1994;161(3):252-7. doi: 10.1007/BF00248701. Arch Microbiol. 1994. PMID: 8161285
-
Biology and taxonomy of Bacillus cereus, Bacillus anthracis, and Bacillus thuringiensis.Can J Microbiol. 2007 Jun;53(6):673-87. doi: 10.1139/W07-029. Can J Microbiol. 2007. PMID: 17668027 Review.
-
The diversity of conjugative relaxases and its application in plasmid classification.FEMS Microbiol Rev. 2009 May;33(3):657-87. doi: 10.1111/j.1574-6976.2009.00168.x. FEMS Microbiol Rev. 2009. PMID: 19396961 Review.
Cited by
-
Highly Efficient Genome Engineering in Bacillus anthracis and Bacillus cereus Using the CRISPR/Cas9 System.Front Microbiol. 2019 Aug 27;10:1932. doi: 10.3389/fmicb.2019.01932. eCollection 2019. Front Microbiol. 2019. PMID: 31551942 Free PMC article.
-
Deeplasmid: deep learning accurately separates plasmids from bacterial chromosomes.Nucleic Acids Res. 2022 Feb 22;50(3):e17. doi: 10.1093/nar/gkab1115. Nucleic Acids Res. 2022. PMID: 34871418 Free PMC article.
References
-
- Guinebretiere MH, Auger S, Galleron N, Contzen M, De Sarrau B, De Buyser ML, Lamberet G, Fagerlund A, Granum PE, Lereclus D, et al. Bacillus cytotoxicus sp. nov. is a novel thermotolerant species of the Bacillus cereus group occasionally associated with food poisoning. Int J Syst Evol Microbiol. 2013;63:31–40. doi: 10.1099/ijs.0.030627-0. - DOI - PubMed
-
- Jimenez G, Urdiain M, Cifuentes A, Lopez-Lopez A, Blanch AR, Tamames J, Kampfer P, Kolsto AB, Ramon D, Martinez JF, et al. Description of Bacillus toyonensis sp. nov., a novel species of the Bacillus cereus group, and pairwise genome comparisons of the species of the group by means of ANI calculations. Syst Appl Microbiol. 2013;36:383–391. doi: 10.1016/j.syapm.2013.04.008. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

