Sequence Polymorphisms at the REDUCED DORMANCY5 Pseudophosphatase Underlie Natural Variation in Arabidopsis Dormancy
- PMID: 27288362
- PMCID: PMC4972279
- DOI: 10.1104/pp.16.00525
Sequence Polymorphisms at the REDUCED DORMANCY5 Pseudophosphatase Underlie Natural Variation in Arabidopsis Dormancy
Abstract
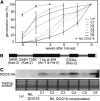
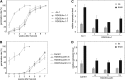
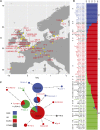
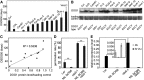
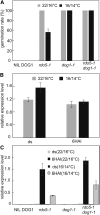
Seed dormancy controls the timing of germination, which regulates the adaptation of plants to their environment and influences agricultural production. The time of germination is under strong natural selection and shows variation within species due to local adaptation. The identification of genes underlying dormancy quantitative trait loci is a major scientific challenge, which is relevant for agricultural and ecological goals. In this study, we describe the identification of the DELAY OF GERMINATION18 (DOG18) quantitative trait locus, which was identified as a factor in natural variation for seed dormancy in Arabidopsis (Arabidopsis thaliana). DOG18 encodes a member of the clade A of the type 2C protein phosphatases family, which we previously identified as the REDUCED DORMANCY5 (RDO5) gene. DOG18/RDO5 shows a relatively high frequency of loss-of-function alleles in natural accessions restricted to northwestern Europe. The loss of dormancy in these loss-of-function alleles can be compensated for by genetic factors like DOG1 and DOG6, and by environmental factors such as low temperature. RDO5 does not have detectable phosphatase activity. Analysis of the phosphoproteome in dry and imbibed seeds revealed a general decrease in protein phosphorylation during seed imbibition that is enhanced in the rdo5 mutant. We conclude that RDO5 acts as a pseudophosphatase that inhibits dephosphorylation during seed imbibition.
© 2016 American Society of Plant Biologists. All Rights Reserved.
Figures

Similar articles
-
Reduced Dormancy5 encodes a protein phosphatase 2C that is required for seed dormancy in Arabidopsis.Plant Cell. 2014 Nov;26(11):4362-75. doi: 10.1105/tpc.114.132811. Epub 2014 Nov 18. Plant Cell. 2014. PMID: 25415980 Free PMC article.
-
The IBO germination quantitative trait locus encodes a phosphatase 2C-related variant with a nonsynonymous amino acid change that interferes with abscisic acid signaling.New Phytol. 2015 Feb;205(3):1076-1082. doi: 10.1111/nph.13225. Epub 2014 Dec 9. New Phytol. 2015. PMID: 25490966
-
Combining association mapping and transcriptomics identify HD2B histone deacetylase as a genetic factor associated with seed dormancy in Arabidopsis thaliana.Plant J. 2013 Jun;74(5):815-28. doi: 10.1111/tpj.12167. Epub 2013 Apr 4. Plant J. 2013. PMID: 23464703
-
The Genetics Underlying Natural Variation in the Biotic Interactions of Arabidopsis thaliana: The Challenges of Linking Evolutionary Genetics and Community Ecology.Curr Top Dev Biol. 2016;119:111-56. doi: 10.1016/bs.ctdb.2016.03.001. Epub 2016 Apr 23. Curr Top Dev Biol. 2016. PMID: 27282025 Review.
-
Plant Ionomics: From Elemental Profiling to Environmental Adaptation.Mol Plant. 2016 Jun 6;9(6):787-97. doi: 10.1016/j.molp.2016.05.003. Epub 2016 May 19. Mol Plant. 2016. PMID: 27212388 Review.
Cited by
-
Seed dormancy release accelerated by elevated partial pressure of oxygen is associated with DOG loci.J Exp Bot. 2018 Jun 27;69(15):3601-3608. doi: 10.1093/jxb/ery156. J Exp Bot. 2018. PMID: 29701795 Free PMC article.
-
Multiple alleles at a single locus control seed dormancy in Swedish Arabidopsis.Elife. 2016 Dec 14;5:e22502. doi: 10.7554/eLife.22502. Elife. 2016. PMID: 27966430 Free PMC article.
-
A commitment for life: Decades of unraveling the molecular mechanisms behind seed dormancy and germination.Plant Cell. 2024 May 1;36(5):1358-1376. doi: 10.1093/plcell/koad328. Plant Cell. 2024. PMID: 38215009 Free PMC article. Review.
-
Transcriptome and proteome analyses reveal the potential mechanism of seed dormancy release in Amomum tsaoko during warm stratification.BMC Genomics. 2023 Mar 2;24(1):99. doi: 10.1186/s12864-023-09202-x. BMC Genomics. 2023. PMID: 36864423 Free PMC article.
-
In Vivo Phosphorylation of the Cytosolic Glucose-6-Phosphate Dehydrogenase Isozyme G6PD6 in Phosphate-Resupplied Arabidopsis thaliana Suspension Cells and Seedlings.Plants (Basel). 2023 Dec 21;13(1):31. doi: 10.3390/plants13010031. Plants (Basel). 2023. PMID: 38202338 Free PMC article.
References
-
- Amiguet-Vercher A, Santuari L, Gonzalez-Guzman M, Depuydt S, Rodriguez PL, Hardtke CS (2015) The IBO germination quantitative trait locus encodes a phosphatase 2C-related variant with a nonsynonymous amino acid change that interferes with abscisic acid signaling. New Phytol 205: 1076–1082 - PubMed
-
- Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 - PubMed
-
- Ashikawa I, Abe F, Nakamura S (2010) Ectopic expression of wheat and barley DOG1-like genes promotes seed dormancy in Arabidopsis. Plant Sci 179: 536–542 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

