The Bradyrhizobium japonicum Ferrous Iron Transporter FeoAB Is Required for Ferric Iron Utilization in Free Living Aerobic Cells and for Symbiosis
- PMID: 27288412
- PMCID: PMC4957049
- DOI: 10.1074/jbc.M116.734129
The Bradyrhizobium japonicum Ferrous Iron Transporter FeoAB Is Required for Ferric Iron Utilization in Free Living Aerobic Cells and for Symbiosis
Abstract
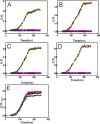
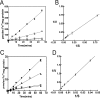
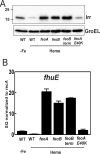
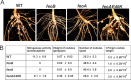
The bacterium Bradyrhizobium japonicum USDA110 does not synthesize siderophores for iron utilization in aerobic environments, and the mechanism of iron uptake within symbiotic soybean root nodules is unknown. An mbfA bfr double mutant defective in iron export and storage activities cannot grow aerobically in very high iron medium. Here, we found that this phenotype was suppressed by loss of function mutations in the feoAB operon encoding ferrous (Fe(2+)) iron uptake proteins. Expression of the feoAB operon genes was elevated under iron limitation, but mutants defective in either gene were unable to grow aerobically over a wide external ferric (Fe(3+)) iron (FeCl3) concentration range. Thus, FeoAB accommodates iron acquisition under iron limited and iron replete conditions. Incorporation of radiolabel from either (55)Fe(2+) or (59)Fe(3+) into cells was severely defective in the feoA and feoB strains, suggesting Fe(3+) reduction to Fe(2+) prior to traversal across the cytoplasmic membrane by FeoAB. The feoA or feoB deletion strains elicited small, ineffective nodules on soybean roots, containing few bacteria and lacking nitrogen fixation activity. A feoA(E40K) mutant contained partial iron uptake activity in culture that supported normal growth and established an effective symbiosis. The feoA(E40K) strain had partial iron uptake activity in situ within nodules and in isolated cells, indicating that FeoAB is the iron transporter in symbiosis. We conclude that FeoAB supports iron acquisition under limited conditions of soil and in the iron-rich environment of a symbiotic nodule.
Keywords: BRADYRHIZOBIUM; bacteria; iron; nitrogen fixation; symbiosis; transport metal.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


Similar articles
-
An Alkane Sulfonate Monooxygenase Is Required for Symbiotic Nitrogen Fixation by Bradyrhizobium diazoefficiens (syn. Bradyrhizobium japonicum) USDA110T.Appl Environ Microbiol. 2019 Nov 27;85(24):e01552-19. doi: 10.1128/AEM.01552-19. Print 2019 Dec 15. Appl Environ Microbiol. 2019. PMID: 31562172 Free PMC article.
-
An iron uptake operon required for proper nodule development in the Bradyrhizobium japonicum-soybean symbiosis.Mol Plant Microbe Interact. 2005 Sep;18(9):950-9. doi: 10.1094/MPMI-18-0950. Mol Plant Microbe Interact. 2005. PMID: 16167765
-
Synthetic Lethality of the bfr and mbfA Genes Reveals a Functional Relationship between Iron Storage and Iron Export in Managing Stress Responses in Bradyrhizobium japonicum.PLoS One. 2016 Jun 10;11(6):e0157250. doi: 10.1371/journal.pone.0157250. eCollection 2016. PLoS One. 2016. PMID: 27285822 Free PMC article.
-
Bacterial ferrous iron transport: the Feo system.FEMS Microbiol Rev. 2016 Mar;40(2):273-98. doi: 10.1093/femsre/fuv049. Epub 2015 Dec 17. FEMS Microbiol Rev. 2016. PMID: 26684538 Review.
-
Iron: an essential micronutrient for the legume-rhizobium symbiosis.Front Plant Sci. 2013 Sep 13;4:359. doi: 10.3389/fpls.2013.00359. Front Plant Sci. 2013. PMID: 24062758 Free PMC article. Review.
Cited by
-
Inter-phylum negative interactions affect soil bacterial community dynamics and functions during soybean development under long-term nitrogen fertilization.Stress Biol. 2021 Nov 26;1(1):15. doi: 10.1007/s44154-021-00015-0. Stress Biol. 2021. PMID: 37676329 Free PMC article.
-
A haem-sequestering plant peptide promotes iron uptake in symbiotic bacteria.Nat Microbiol. 2022 Sep;7(9):1453-1465. doi: 10.1038/s41564-022-01192-y. Epub 2022 Aug 11. Nat Microbiol. 2022. PMID: 35953657 Free PMC article.
-
The Medicago truncatula Vacuolar iron Transporter-Like proteins VTL4 and VTL8 deliver iron to symbiotic bacteria at different stages of the infection process.New Phytol. 2020 Oct;228(2):651-666. doi: 10.1111/nph.16735. Epub 2020 Jul 16. New Phytol. 2020. PMID: 32521047 Free PMC article.
-
Prokaryotic Heme Biosynthesis: Multiple Pathways to a Common Essential Product.Microbiol Mol Biol Rev. 2017 Jan 25;81(1):e00048-16. doi: 10.1128/MMBR.00048-16. Print 2017 Mar. Microbiol Mol Biol Rev. 2017. PMID: 28123057 Free PMC article. Review.
-
Disentangling the Evolutionary History of Feo, the Major Ferrous Iron Transport System in Bacteria.mBio. 2022 Feb 22;13(1):e0351221. doi: 10.1128/mbio.03512-21. Epub 2022 Jan 11. mBio. 2022. PMID: 35012344 Free PMC article.
References
-
- Andrews S. C., Robinson A. K., and Rodríguez-Quiñones F. (2003) Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237 - PubMed
-
- Chu B. C., Garcia-Herrero A., Johanson T. H., Krewulak K. D., Lau C. K., Peacock R. S., Slavinskaya Z., and Vogel H. J. (2010) Siderophore uptake in bacteria and the battle for iron with the host: a bird's eye view. Biometals 23, 601–611 - PubMed
-
- Faraldo-Gómez J. D., and Sansom M. S. (2003) Acquisition of siderophores in Gram-negative bacteria. Nat. Rev. Mol. Cell Biol. 4, 105–116 - PubMed
-
- Wandersman C., and Stojiljkovic I. (2000) Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr. Opin. Microbiol. 3, 215–220 - PubMed
-
- Genco C. A., and Dixon D. W. (2001) Emerging strategies in microbial haem capture. Mol. Microbiol. 39, 1–11 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

