Mechanisms of transcription factor evolution in Metazoa
- PMID: 27288445
- PMCID: PMC5291267
- DOI: 10.1093/nar/gkw492
Mechanisms of transcription factor evolution in Metazoa
Abstract
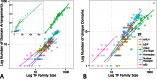
Transcriptions factors (TFs) are pivotal for the regulation of virtually all cellular processes, including growth and development. Expansions of TF families are causally linked to increases in organismal complexity. Here we study the evolutionary dynamics, genetic causes and functional implications of the five largest metazoan TF families. We find that family expansions dominate across the whole metazoan tree; however, some branches experience exceptional family-specific accelerated expansions. Additionally, we find that such expansions are often predated by modular domain rearrangements, which spur the expansion of a new sub-family by separating it from the rest of the TF family in terms of protein-protein interactions. This separation allows for radical shifts in the functional spectrum of a duplicated TF. We also find functional differentiation inside TF sub-families as changes in expression specificity. Furthermore, accelerated family expansions are facilitated by repeats of sequence motifs such as C2H2 zinc fingers. We quantify whole genome duplications and single gene duplications as sources of TF family expansions, implying that some, but not all, TF duplicates are preferentially retained. We conclude that trans-regulatory changes (domain rearrangements) are instrumental for fundamental functional innovations, that cis-regulatory changes (affecting expression) accomplish wide-spread fine tuning and both jointly contribute to the functional diversification of TFs.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Valentine J.W., Collins A.G., Meyer C.P. Morphological complexity increase in metazoans. Paleobiology. 1994;20:131–142.
-
- Degnan B.M., Vervoort M., Larroux C., Richards G.S. Early evolution of metazoan transcription factors. Curr. Opin. Genet. Dev. 2009;19:591–599. - PubMed
-
- Miyata T., Suga H. Divergence pattern of animal gene families and relationship with the Cambrian explosion. Bioessays. 2001;23:1018–1027. - PubMed
-
- Levine M., Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

