RTFBSDB: an integrated framework for transcription factor binding site analysis
- PMID: 27288497
- PMCID: PMC5039921
- DOI: 10.1093/bioinformatics/btw338
RTFBSDB: an integrated framework for transcription factor binding site analysis
Abstract
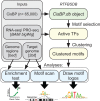
Transcription factors (TFs) regulate complex programs of gene transcription by binding to short DNA sequence motifs. Here, we introduce rtfbsdb, a unified framework that integrates a database of more than 65 000 TF binding motifs with tools to easily and efficiently scan target genome sequences. Rtfbsdb clusters motifs with similar DNA sequence specificities and integrates RNA-seq or PRO-seq data to restrict analyses to motifs recognized by TFs expressed in the cell type of interest. Our package allows common analyses to be performed rapidly in an integrated environment.
Availability and implementation: rtfbsdb available at (https://github.com/Danko-Lab/rtfbs_db).
Contact: dankoc@gmail.com
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2016. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

