Independent Domestication of Two Old World Cotton Species
- PMID: 27289095
- PMCID: PMC4943200
- DOI: 10.1093/gbe/evw129
Independent Domestication of Two Old World Cotton Species
Abstract
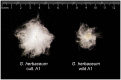
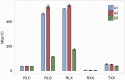
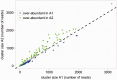
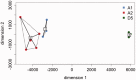
Domesticated cotton species provide raw material for the majority of the world's textile industry. Two independent domestication events have been identified in allopolyploid cotton, one in Upland cotton (Gossypium hirsutum L.) and the other to Egyptian cotton (Gossypium barbadense L.). However, two diploid cotton species, Gossypium arboreum L. and Gossypium herbaceum L., have been cultivated for several millennia, but their status as independent domesticates has long been in question. Using genome resequencing data, we estimated the global abundance of various repetitive DNAs. We demonstrate that, despite negligible divergence in genome size, the two domesticated diploid cotton species contain different, but compensatory, repeat content and have thus experienced cryptic alterations in repeat abundance despite equivalence in genome size. Evidence of independent origin is bolstered by estimates of divergence times based on molecular evolutionary analysis of f7,000 orthologous genes, for which synonymous substitution rates suggest that G. arboreum and G. herbaceum last shared a common ancestor approximately 0.4-2.5 Ma. These data are incompatible with a shared domestication history during the emergence of agriculture and lead to the conclusion that G. arboreum and G. herbaceum were each domesticated independently.
Keywords: Gossypium; crop plants; genome size; molecular evolution; repetitive DNA.
© The Author(s) 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
Similar articles
-
Parallel expression evolution of oxidative stress-related genes in fiber from wild and domesticated diploid and polyploid cotton (Gossypium).BMC Genomics. 2009 Aug 17;10:378. doi: 10.1186/1471-2164-10-378. BMC Genomics. 2009. PMID: 19686594 Free PMC article.
-
The Gossypium herbaceum L. Wagad genome as a resource for understanding cotton domestication.G3 (Bethesda). 2023 Feb 9;13(2):jkac308. doi: 10.1093/g3journal/jkac308. G3 (Bethesda). 2023. PMID: 36454094 Free PMC article.
-
Parallel and Intertwining Threads of Domestication in Allopolyploid Cotton.Adv Sci (Weinh). 2021 Mar 15;8(10):2003634. doi: 10.1002/advs.202003634. eCollection 2021 May. Adv Sci (Weinh). 2021. PMID: 34026441 Free PMC article.
-
Dual Domestication, Diversity, and Differential Introgression in Old World Cotton Diploids.Genome Biol Evol. 2022 Dec 7;14(12):evac170. doi: 10.1093/gbe/evac170. Genome Biol Evol. 2022. PMID: 36510772 Free PMC article.
-
[A and D genome evolution in Gossypium revealed using SSR molecular markers].Yi Chuan Xue Bao. 2003 Feb;30(2):183-8. Yi Chuan Xue Bao. 2003. PMID: 12776608 Chinese.
Cited by
-
Genome-Wide Identification and Characterization of CPR5 Genes in Gossypium Reveals Their Potential Role in Trichome Development.Front Genet. 2022 Jun 8;13:921096. doi: 10.3389/fgene.2022.921096. eCollection 2022. Front Genet. 2022. PMID: 35754813 Free PMC article.
-
Features of Chromosome Introgression from Gossypium barbadense L. into G. hirsutum L. during the Development of Alien Substitution Lines.Plants (Basel). 2022 Feb 18;11(4):542. doi: 10.3390/plants11040542. Plants (Basel). 2022. PMID: 35214875 Free PMC article.
-
Genomic analyses reveal the genetic basis of early maturity and identification of loci and candidate genes in upland cotton (Gossypium hirsutum L.).Plant Biotechnol J. 2021 Jan;19(1):109-123. doi: 10.1111/pbi.13446. Epub 2020 Aug 1. Plant Biotechnol J. 2021. PMID: 32652678 Free PMC article.
-
Nucleotide Evolution, Domestication Selection, and Genetic Relationships of Chloroplast Genomes in the Economically Important Crop Genus Gossypium.Front Plant Sci. 2022 Apr 15;13:873788. doi: 10.3389/fpls.2022.873788. eCollection 2022. Front Plant Sci. 2022. PMID: 35498673 Free PMC article.
-
Genome-Wide Mining of MYB Transcription Factors in the Anthocyanin Biosynthesis Pathway of Gossypium Hirsutum.Biochem Genet. 2021 Jun;59(3):678-696. doi: 10.1007/s10528-021-10027-0. Epub 2021 Jan 27. Biochem Genet. 2021. PMID: 33502632
References
-
- Abbo S, Lev-Yadun S, Gopher A. 2012. Plant domestication and crop evolution in the Near East: on events and processes. Crit Rev Plant Sci. 31:241–257.
-
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate–a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 57:289–300.
-
- Bennett MD, Leitch IJ. 2010. Angiosperm DNA C-values database. Available from: http://data.kew.org/cvalues/.
-
- Brubaker C, Paterson A, Wendel J. 1999. Comparative genetic mapping of allotetraploid cotton and its diploid progenitors. Genome 42:184–203.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

