GhERF-IIb3 regulates the accumulation of jasmonate and leads to enhanced cotton resistance to blight disease
- PMID: 27291786
- PMCID: PMC6638235
- DOI: 10.1111/mpp.12445
GhERF-IIb3 regulates the accumulation of jasmonate and leads to enhanced cotton resistance to blight disease
Abstract
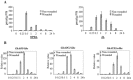
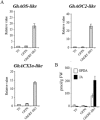
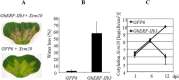
The phytohormone jasmonic acid (JA) and its derivatives, collectively referred to as jasmonates, regulate many developmental processes, but are also involved in the response to numerous abiotic/biotic stresses. Thus far, powerful reverse genetic strategies employing perception, signalling or biosynthesis mutants have broadly contributed to our understanding of the role of JA in the plant stress response and development, as has the chemical gain-of-function approach based on exogenous application of the hormone. However, there is currently no method that allows for tightly controlled JA production in planta. By investigating the control of the JA synthesis pathway in bacteria-infected cotton (Gossypium hirsutum L.) plants, we identified a transcription factor (TF), named GhERF-IIb3, which acts as a positive regulator of the JA pathway. Expression of this well-conserved TF in cotton leaves was sufficient to produce in situ JA accumulation at physiological concentrations associated with an enhanced cotton defence response to bacterial infection.
Keywords: ERF transcription factor; cotton; defence response; jasmonate, Xanthomonas citri pv. malvacearum.
© 2016 BSPP AND JOHN WILEY & SONS LTD.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures



References
-
- Browse, J. (2009) Jasmonate passes muster: a receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60, 183–205. - PubMed
-
- Cacas, J.L. , Marmey, P. , Montillet, J.L. , Sayegh‐Alhamdia, M. , Jalloul, A. , Rojas‐Mendoza, A. , Clérivet, A. and Nicole, M. (2009) A novel patatin‐like protein from cotton plant, GhPat1, is co‐expressed with GhLox1 during Xanthomonas campestris‐mediated hypersensitive cell death. Plant Cell Rep. 28, 155–164. - PubMed
-
- Champion, A. , Hebrard, E. , Parra, B. , Bournaud, C. , Marmey, P. , Tranchant, C. and Nicole, M. (2009) Molecular diversity and gene expression of cotton ERF transcription factors reveal that group IXa members are responsive to jasmonate, ethylene and Xanthomonas . Mol. Plant Pathol. 10, 471–485. - PMC - PubMed
-
- Cui, H. , Tsuda, K. and Parker, J.E. (2015) Effector‐triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

