Biallelic Mutations of VAC14 in Pediatric-Onset Neurological Disease
- PMID: 27292112
- PMCID: PMC5005439
- DOI: 10.1016/j.ajhg.2016.05.008
Biallelic Mutations of VAC14 in Pediatric-Onset Neurological Disease
Abstract
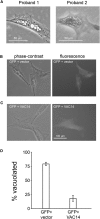
In the PI(3,5)P2 biosynthetic complex, the lipid kinase PIKFYVE and the phosphatase FIG4 are bound to the dimeric scaffold protein VAC14, which is composed of multiple heat-repeat domains. Mutations of FIG4 result in the inherited disorders Charcot-Marie-Tooth disease type 4J, Yunis-Varón syndrome, and polymicrogyria with seizures. We here describe inherited variants of VAC14 in two unrelated children with sudden onset of a progressive neurological disorder and regression of developmental milestones. Both children developed impaired movement with dystonia, became nonambulatory and nonverbal, and exhibited striatal abnormalities on MRI. A diagnosis of Leigh syndrome was rejected due to normal lactate profiles. Exome sequencing identified biallelic variants of VAC14 that were inherited from unaffected heterozygous parents in both families. Proband 1 inherited a splice-site variant that results in skipping of exon 13, p.Ile459Profs(∗)4 (not reported in public databases), and the missense variant p.Trp424Leu (reported in the ExAC database in a single heterozygote). Proband 2 inherited two missense variants in the dimerization domain of VAC14, p.Ala582Ser and p.Ser583Leu, that have not been previously reported. Cultured skin fibroblasts exhibited the accumulation of vacuoles that is characteristic of PI(3,5)P2 deficiency. Vacuolization of fibroblasts was rescued by transfection of wild-type VAC14 cDNA. The similar age of onset and neurological decline in the two unrelated children define a recessive disorder resulting from compound heterozygosity for deleterious variants of VAC14.
Copyright © 2016 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Cooke F.T. Phosphatidylinositol 3,5-bisphosphate: metabolism and function. Arch. Biochem. Biophys. 2002;407:143–151. - PubMed
-
- Dove S.K., Dong K., Kobayashi T., Williams F.K., Michell R.H. Phosphatidylinositol 3,5-bisphosphate and Fab1p/PIKfyve underPPIn endo-lysosome function. Biochem. J. 2009;419:1–13. - PubMed
-
- Ho C.Y., Alghamdi T.A., Botelho R.J. Phosphatidylinositol-3,5-bisphosphate: no longer the poor PIP2. Traffic. 2012;13:1–8. - PubMed
-
- Michell R.H. Inositol lipids: from an archaeal origin to phosphatidylinositol 3,5-bisphosphate faults in human disease. FEBS J. 2013;280:6281–6294. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

