PiggyBac transposon-based polyadenylation-signal trap for genome-wide mutagenesis in mice
- PMID: 27292714
- PMCID: PMC4904408
- DOI: 10.1038/srep27788
PiggyBac transposon-based polyadenylation-signal trap for genome-wide mutagenesis in mice
Abstract
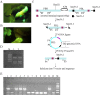
We designed a new type of polyadenylation-signal (PAS) trap vector system in living mice, the piggyBac (PB) (PAS-trapping (EGFP)) gene trapping vector, which takes advantage of the efficient transposition ability of PB and efficient gene trap and insertional mutagenesis of PAS-trapping. The reporter gene of PB(PAS-trapping (EGFP)) is an EGFP gene with its own promoter, but lacking a poly(A) signal. Transgenic mouse lines carrying PB(PAS-trapping (EGFP)) and protamine 1 (Prm1) promoter-driven PB transposase transgenes (Prm1-PBase) were generated by microinjection. Male mice doubly positive for PB(PAS-trapping (EGFP)) and Prm1-PBase were crossed with WT females, generating offspring with various insertion mutations. We found that 44.8% (26/58) of pups were transposon-positive progenies. New transposon integrations comprised 26.9% (7/26) of the transposon-positive progenies. We found that 100% (5/5) of the EGFP fluorescence-positive mice had new trap insertions mediated by a PB transposon in transcriptional units. The direction of the EGFP gene in the vector was consistent with the direction of the endogenous gene reading frame. Furthermore, mice that were EGFP-PCR positive, but EGFP fluorescent negative, did not show successful gene trapping. Thus, the novel PB(PAS-trapping (EGFP)) system is an efficient genome-wide gene-trap mutagenesis in mice.
Figures



Similar articles
-
Insertional mutagenesis by a hybrid piggyBac and sleeping beauty transposon in the rat.Genetics. 2012 Dec;192(4):1235-48. doi: 10.1534/genetics.112.140855. Epub 2012 Sep 28. Genetics. 2012. PMID: 23023007 Free PMC article.
-
Generating mutant rats using the Sleeping Beauty transposon system.Methods. 2009 Nov;49(3):236-42. doi: 10.1016/j.ymeth.2009.04.010. Epub 2009 May 4. Methods. 2009. PMID: 19398007
-
Transposon mutagenesis of the mouse germline.Genetics. 2003 Sep;165(1):243-56. doi: 10.1093/genetics/165.1.243. Genetics. 2003. PMID: 14504232 Free PMC article.
-
[The improvement and application of piggyBac transposon system in mammals].Yi Chuan. 2014 Oct;36(10):965-73. doi: 10.3724/SP.J.1005.2014.0965. Yi Chuan. 2014. PMID: 25406244 Review. Chinese.
-
Sleeping Beauty transposon insertional mutagenesis based mouse models for cancer gene discovery.Curr Opin Genet Dev. 2015 Feb;30:66-72. doi: 10.1016/j.gde.2015.04.007. Epub 2015 Jun 4. Curr Opin Genet Dev. 2015. PMID: 26051241 Free PMC article. Review.
Cited by
-
Ablation of Zfhx4 results in early postnatal lethality by disrupting the respiratory center in mice.J Mol Cell Biol. 2021 Jul 6;13(3):210-224. doi: 10.1093/jmcb/mjaa081. J Mol Cell Biol. 2021. PMID: 33475140 Free PMC article.
-
Giving the Genes a Shuffle: Using Natural Variation to Understand Host Genetic Contributions to Viral Infections.Trends Genet. 2018 Oct;34(10):777-789. doi: 10.1016/j.tig.2018.07.005. Epub 2018 Aug 18. Trends Genet. 2018. PMID: 30131185 Free PMC article. Review.
-
Identification of genes associated with cortical malformation using a transposon-mediated somatic mutagenesis screen in mice.Nat Commun. 2018 Jun 27;9(1):2498. doi: 10.1038/s41467-018-04880-8. Nat Commun. 2018. PMID: 29950674 Free PMC article.
References
-
- Lander E. S. et al.. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). - PubMed
-
- Venter J. C. et al.. The sequence of the human genome. Science 291, 1304–1351 (2001). - PubMed
-
- Finishing the euchromatic sequence of the human genome. Nature 431, 931–945 (2004). - PubMed
-
- Lander E. S. Initial impact of the sequencing of the human genome. Nature 470, 187–197 (2011). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

