Cutoff Suppresses RNA Polymerase II Termination to Ensure Expression of piRNA Precursors
- PMID: 27292797
- PMCID: PMC4980073
- DOI: 10.1016/j.molcel.2016.05.010
Cutoff Suppresses RNA Polymerase II Termination to Ensure Expression of piRNA Precursors
Abstract
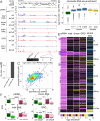
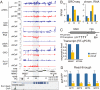
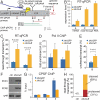
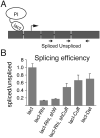
Small non-coding RNAs called piRNAs serve as guides for an adaptable immune system that represses transposable elements in germ cells of Metazoa. In Drosophila the RDC complex, composed of Rhino, Deadlock and Cutoff (Cuff) bind chromatin of dual-strand piRNA clusters, special genomic regions, which encode piRNA precursors. The RDC complex is required for transcription of piRNA precursors, though the mechanism by which it licenses transcription remained unknown. Here, we show that Cuff prevents premature termination of RNA polymerase II. Cuff prevents cleavage of nascent RNA at poly(A) sites by interfering with recruitment of the cleavage and polyadenylation specificity factor (CPSF) complex. Cuff also protects processed transcripts from degradation by the exonuclease Rat1. Our work reveals a conceptually different mechanism of transcriptional enhancement. In contrast to other factors that regulate termination by binding to specific signals on nascent RNA, the RDC complex inhibits termination in a chromatin-dependent and sequence-independent manner.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures



References
-
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. - PubMed
-
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
