Mitochondrial Dynamics Controls T Cell Fate through Metabolic Programming
- PMID: 27293185
- PMCID: PMC4974356
- DOI: 10.1016/j.cell.2016.05.035
Mitochondrial Dynamics Controls T Cell Fate through Metabolic Programming
Abstract
Activated effector T (TE) cells augment anabolic pathways of metabolism, such as aerobic glycolysis, while memory T (TM) cells engage catabolic pathways, like fatty acid oxidation (FAO). However, signals that drive these differences remain unclear. Mitochondria are metabolic organelles that actively transform their ultrastructure. Therefore, we questioned whether mitochondrial dynamics controls T cell metabolism. We show that TE cells have punctate mitochondria, while TM cells maintain fused networks. The fusion protein Opa1 is required for TM, but not TE cells after infection, and enforcing fusion in TE cells imposes TM cell characteristics and enhances antitumor function. Our data suggest that, by altering cristae morphology, fusion in TM cells configures electron transport chain (ETC) complex associations favoring oxidative phosphorylation (OXPHOS) and FAO, while fission in TE cells leads to cristae expansion, reducing ETC efficiency and promoting aerobic glycolysis. Thus, mitochondrial remodeling is a signaling mechanism that instructs T cell metabolic programming.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
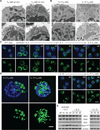


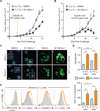


Comment in
-
T cells: Mitochondrial shape shifters.Nat Rev Immunol. 2016 Jun 28;16(7):402-3. doi: 10.1038/nri.2016.79. Nat Rev Immunol. 2016. PMID: 27349280 No abstract available.
-
Mitochondrial Networking in T Cell Memory.Cell. 2016 Jun 30;166(1):9-10. doi: 10.1016/j.cell.2016.06.035. Cell. 2016. PMID: 27368094
-
Mitochondrial fusion fuels T cell memory.Cell Res. 2016 Sep;26(9):969-70. doi: 10.1038/cr.2016.94. Epub 2016 Aug 12. Cell Res. 2016. PMID: 27514702 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

