Forces Driving Chaperone Action
- PMID: 27293188
- PMCID: PMC4947014
- DOI: 10.1016/j.cell.2016.05.054
Forces Driving Chaperone Action
Abstract
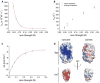
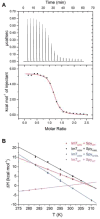
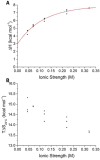
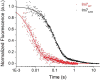
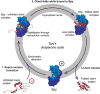
It is still unclear what molecular forces drive chaperone-mediated protein folding. Here, we obtain a detailed mechanistic understanding of the forces that dictate the four key steps of chaperone-client interaction: initial binding, complex stabilization, folding, and release. Contrary to the common belief that chaperones recognize unfolding intermediates by their hydrophobic nature, we discover that the model chaperone Spy uses long-range electrostatic interactions to rapidly bind to its unfolded client protein Im7. Short-range hydrophobic interactions follow, which serve to stabilize the complex. Hydrophobic collapse of the client protein then drives its folding. By burying hydrophobic residues in its core, the client's affinity to Spy decreases, which causes client release. By allowing the client to fold itself, Spy circumvents the need for client-specific folding instructions. This mechanism might help explain how chaperones can facilitate the folding of various unrelated proteins.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures

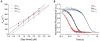
Similar articles
-
Increased surface charge in the protein chaperone Spy enhances its anti-aggregation activity.J Biol Chem. 2020 Oct 16;295(42):14488-14500. doi: 10.1074/jbc.RA119.012300. Epub 2020 Aug 17. J Biol Chem. 2020. PMID: 32817055 Free PMC article.
-
Electrostatic interactions are important for chaperone-client interaction in vivo.Microbiology (Reading). 2018 Jul;164(7):992-997. doi: 10.1099/mic.0.000676. Epub 2018 Jun 5. Microbiology (Reading). 2018. PMID: 29870331 Free PMC article.
-
A molecular mechanism of chaperone-client recognition.Sci Adv. 2016 Nov 16;2(11):e1601625. doi: 10.1126/sciadv.1601625. eCollection 2016 Nov. Sci Adv. 2016. PMID: 28138538 Free PMC article.
-
Chaperone-client interactions: Non-specificity engenders multifunctionality.J Biol Chem. 2017 Jul 21;292(29):12010-12017. doi: 10.1074/jbc.R117.796862. Epub 2017 Jun 15. J Biol Chem. 2017. PMID: 28620048 Free PMC article. Review.
-
Folding while bound to chaperones.Curr Opin Struct Biol. 2018 Feb;48:1-5. doi: 10.1016/j.sbi.2017.06.009. Epub 2017 Jul 19. Curr Opin Struct Biol. 2018. PMID: 28734135 Free PMC article. Review.
Cited by
-
Why? - Successful Pseudomonas aeruginosa clones with a focus on clone C.FEMS Microbiol Rev. 2020 Nov 24;44(6):740-762. doi: 10.1093/femsre/fuaa029. FEMS Microbiol Rev. 2020. PMID: 32990729 Free PMC article. Review.
-
Molecular chaperones: providing a safe place to weather a midlife protein-folding crisis.Nat Struct Mol Biol. 2016 Jul 6;23(7):621-3. doi: 10.1038/nsmb.3255. Nat Struct Mol Biol. 2016. PMID: 27384188 No abstract available.
-
Increased surface charge in the protein chaperone Spy enhances its anti-aggregation activity.J Biol Chem. 2020 Oct 16;295(42):14488-14500. doi: 10.1074/jbc.RA119.012300. Epub 2020 Aug 17. J Biol Chem. 2020. PMID: 32817055 Free PMC article.
-
Amyloid management by chaperones: The mystery underlying protein oligomers' dual functions.Curr Res Struct Biol. 2022 Dec 7;4:356-364. doi: 10.1016/j.crstbi.2022.11.002. eCollection 2022. Curr Res Struct Biol. 2022. PMID: 36523328 Free PMC article. Review.
-
An increase in surface hydrophobicity mediates chaperone activity in N-chlorinated RidA.Redox Biol. 2022 Jul;53:102332. doi: 10.1016/j.redox.2022.102332. Epub 2022 May 7. Redox Biol. 2022. PMID: 35598378 Free PMC article.
References
-
- Berg OG, von Hippel PH. Diffusion-controlled macromolecular interactions. Annu Rev Biophys Biophys Chem. 1985;14:131–160. - PubMed
-
- Capaldi AP, Shastry MCR, Kleanthous C, Roder H, Radford SE. Ultrarapid mixing experiments reveal that Im7 folds via an on-pathway intermediate. Nat Struct Biol. 2001;8:68–72. - PubMed
-
- Capaldi AP, Kleanthous C, Radford SE. Im7 folding mechanism: misfolding on a path to the native state. Nat Struct Biol. 2002;9:209–216. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

