Restricted Location of PSEN2/γ-Secretase Determines Substrate Specificity and Generates an Intracellular Aβ Pool
- PMID: 27293189
- PMCID: PMC7439524
- DOI: 10.1016/j.cell.2016.05.020
Restricted Location of PSEN2/γ-Secretase Determines Substrate Specificity and Generates an Intracellular Aβ Pool
Abstract
γ-Secretases are a family of intramembrane-cleaving proteases involved in various signaling pathways and diseases, including Alzheimer's disease (AD). Cells co-express differing γ-secretase complexes, including two homologous presenilins (PSENs). We examined the significance of this heterogeneity and identified a unique motif in PSEN2 that directs this γ-secretase to late endosomes/lysosomes via a phosphorylation-dependent interaction with the AP-1 adaptor complex. Accordingly, PSEN2 selectively cleaves late endosomal/lysosomal localized substrates and generates the prominent pool of intracellular Aβ that contains longer Aβ; familial AD (FAD)-associated mutations in PSEN2 increased the levels of longer Aβ further. Moreover, a subset of FAD mutants in PSEN1, normally more broadly distributed in the cell, phenocopies PSEN2 and shifts its localization to late endosomes/lysosomes. Thus, localization of γ-secretases determines substrate specificity, while FAD-causing mutations strongly enhance accumulation of aggregation-prone Aβ42 in intracellular acidic compartments. The findings reveal potentially important roles for specific intracellular, localized reactions contributing to AD pathogenesis.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures



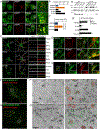


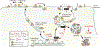
Comment in
-
Sorting Out Presenilins in Alzheimer's Disease.Cell. 2016 Jun 30;166(1):13-5. doi: 10.1016/j.cell.2016.06.034. Cell. 2016. PMID: 27368096
Similar articles
-
Dissociation between the processivity and total activity of γ-secretase: implications for the mechanism of Alzheimer's disease-causing presenilin mutations.Biochemistry. 2011 Oct 25;50(42):9023-35. doi: 10.1021/bi2007146. Epub 2011 Sep 30. Biochemistry. 2011. PMID: 21919498 Free PMC article.
-
Novel presenilin 1 and 2 double knock-out cell line for in vitro validation of PSEN1 and PSEN2 mutations.Neurobiol Dis. 2020 May;138:104785. doi: 10.1016/j.nbd.2020.104785. Epub 2020 Feb 4. Neurobiol Dis. 2020. PMID: 32032730 Free PMC article.
-
Dominant negative effect of the loss-of-function γ-secretase mutants on the wild-type enzyme through heterooligomerization.Proc Natl Acad Sci U S A. 2017 Nov 28;114(48):12731-12736. doi: 10.1073/pnas.1713605114. Epub 2017 Oct 9. Proc Natl Acad Sci U S A. 2017. PMID: 29078389 Free PMC article.
-
Evidence For and Against a Pathogenic Role of Reduced γ-Secretase Activity in Familial Alzheimer's Disease.J Alzheimers Dis. 2016 Apr 4;52(3):781-99. doi: 10.3233/JAD-151186. J Alzheimers Dis. 2016. PMID: 27060961 Review.
-
Toward the structure of presenilin/γ-secretase and presenilin homologs.Biochim Biophys Acta. 2013 Dec;1828(12):2886-97. doi: 10.1016/j.bbamem.2013.04.015. Biochim Biophys Acta. 2013. PMID: 24099007 Free PMC article. Review.
Cited by
-
Probing γ-secretase-substrate interactions at the single amino acid residue level.EMBO J. 2016 Aug 1;35(15):1597-9. doi: 10.15252/embj.201694978. Epub 2016 Jul 1. EMBO J. 2016. PMID: 27370209 Free PMC article.
-
Spatial snapshots of amyloid precursor protein intramembrane processing via early endosome proteomics.Nat Commun. 2022 Oct 16;13(1):6112. doi: 10.1038/s41467-022-33881-x. Nat Commun. 2022. PMID: 36245040 Free PMC article.
-
Analysis of Non-Amyloidogenic Mutations in APP Supports Loss of Function Hypothesis of Alzheimer's Disease.Int J Mol Sci. 2023 Jan 20;24(3):2092. doi: 10.3390/ijms24032092. Int J Mol Sci. 2023. PMID: 36768421 Free PMC article.
-
Presenilin gene function and Notch signaling feedback regulation in the developing mouse lens.Differentiation. 2018 Jul-Aug;102:40-52. doi: 10.1016/j.diff.2018.07.003. Epub 2018 Jul 25. Differentiation. 2018. PMID: 30059908 Free PMC article.
-
Converging roles of PSENEN/PEN2 and CLN3 in the autophagy-lysosome system.Autophagy. 2022 Sep;18(9):2068-2085. doi: 10.1080/15548627.2021.2016232. Epub 2021 Dec 29. Autophagy. 2022. PMID: 34964690 Free PMC article.
References
-
- Bayer TA, and Wirths O (2011). Intraneuronal Abeta as a trigger for neuron loss: can this be translated into human pathology? Biochemical Society transactions 39, 857–861. - PubMed
-
- Bentahir M, Nyabi O, Verhamme J, Tolia A, Horre K, Wiltfang J, Esselmann H, and De Strooper B (2006). Presenilin clinical mutations can affect gamma-secretase activity by different mechanisms. Journal of neurochemistry 96, 732–742. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

